Magnetic Resonance Spectroscopy Deep Learning Denoising Using Few In Vivo Data(中文,English)
Dicheng Chen1,#, Wanqi Hu1,#, Huiting Liu1, Yirong Zhou1,Tianyu Qiu1,Yihui Huang1,Zi Wang1,Meijin Lin2, Liangjie Lin3, Zhigang Wu3, Jiazheng Wang3, Hao Chen4, Xi Chen5, Gen Yan6,Di Guo7,Jianzhong Lin8,Xiaobo Qu1,*
1Department of Electronic Science, Fujian Provincial Key Laboratory of Plasma and Magnetic Resonance, Xiamen University, Xiamen 361005, China.
2Department of Applied Marine Physics & Engineering, College of Ocean and Earth Sciences, Xiamen University, Xiamen 361005, China.
3Philips, Beijing 100016, China.
4School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai 200240, China.
5McLean Hospital, Harvard Medical School, Belmont, MA 02478, USA.
6Department of Radiology, The Second Hospital Affiliated to Xiamen Medical College, Xiamen 361021, China.
7School of Computer and Information Engineering, Xiamen University of Technology, Xiamen 361024, China.
8Department of Radiology, Zhongshan Hospital affiliated to Xiamen University, Xiamen 361004, China.
*Email:quxiaobo <at> xmu.edu.cn
Citation
D. Chen, W. Hu, H. Liu, Y. Zhou, T. Qiu, Y. Huang, Z. Wang, M. Lin, L. Lin, Z. Wu, J. Wang, H. Chen, X. Chen, G. Yan, D. Guo, J. Lin, and X. Qu, Magnetic resonance spectroscopy deep learning denoising using few in vivo data, IEEE Transactions on Computational Imaging, DOI: 10.1109/TCI.2023.3267623, 2023.
Synopsis
Magnetic Resonance Spectroscopy (MRS) is a noninvasive tool to reveal metabolic information. One challenge of 1H-MRS is the low Signal-Noise Ratio (SNR)[1-2]. To improve the SNR, a typical approach is to perform a large Number of Signal Averaging (NSA)[3-4]. The data acquisition time, however, is proportional to the NSA accordingly. A complete clinical MRS scan takes approximately 10 minutes in a common setting with an NSA of 128. Recently, deep learning has been introduced to improve the SNR, but generally, only the simulated data are used as the training set. This may hinder the MRS applications since some non-ideal conditions in practical, such as imperfections of acquisition system, and subjects’ physiological activities or psychologic status, etc., are not considered for the simulation data.
Main Context
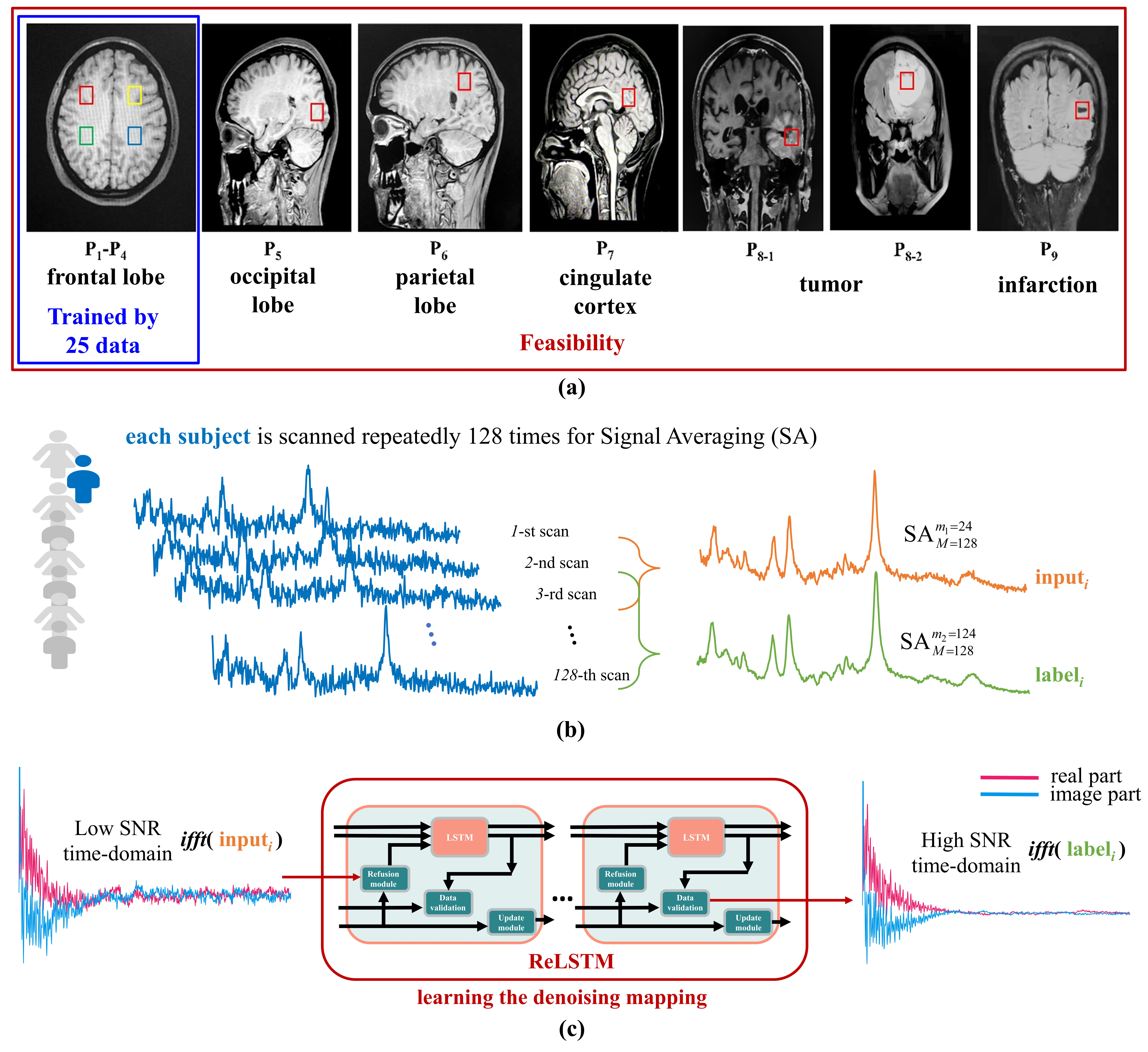
In this paper, a new scheme that purely uses part of repeated samplings of realistic data was proposed, instead of physics-driven synthetic data[5]. A deep learning model, Refusion Long Short-Term Memory (ReLSTM), was designed to learn the mapping from the time-domain data with low SNR (24 NSA) to the one with high SNR (128 NSA). To train this model, we combined different repetitions of single-voxel MRS data of in vivo human brains to construct a substantial realistic training set. A large number of training data can be obtained by randomly selecting m repeated samplings from the M (M > m) repetitions. Following this data augmentation strategy, a few number (25) of healthy frontal lobes spectra were collected as the training sets and each set produces 1000 items for training. For the feasibility test, we chose the spectra not only of frontal lobe (P1-4), but also of other positions, such as occipital lobe, parietal lobe, cingulate cortex (P5-P7), and even lesion region (P8-P9), from different subjects. All of the training steps are shown in Figure 1.

Fig. 1. The entire training phase of the proposed ReLSTM. (a) Data arrangement: 25 spectra of healthy frontal lobes are used to train the ReLSTM, and 4 spectra of frontal lobes, 3 of other regions and 3 of lesion regions are for feasibility test. (b) The generation of one training input: a input is an average of m1=24 repeated samplings which are randomly selected out of the total M=128 repeated scans. (c) The proposed denoising ReLSTM structure. It was trained for fitting the mapping, in the time domain, between the input of low SNR and the corresponding high SNR label, where ifft represents inverse Fourier
Experiments on the in vivo brains of the regions of 7 healthy, 2 tumors and 1 cerebral infarction (total 1000 spectra) show that using NSA of 24, only 20% of a common setting, the spectra denoised by ReLSTM can provide the estimated concentrations of metabolites with the reliability comparable to those of the high-SNR spectra obtained commonly with 128 SA, which are shown in Table 1. Furthermore, compared with the state-of-the-art Low-Rank[6] denoising method, ReLSTM achieves lower relative errors and the Cramér-Rao lower bounds for quantifying some important biomarkers. More details can be accessed in the full paper: DOI: 10.1109/TCI.2023.3267623.
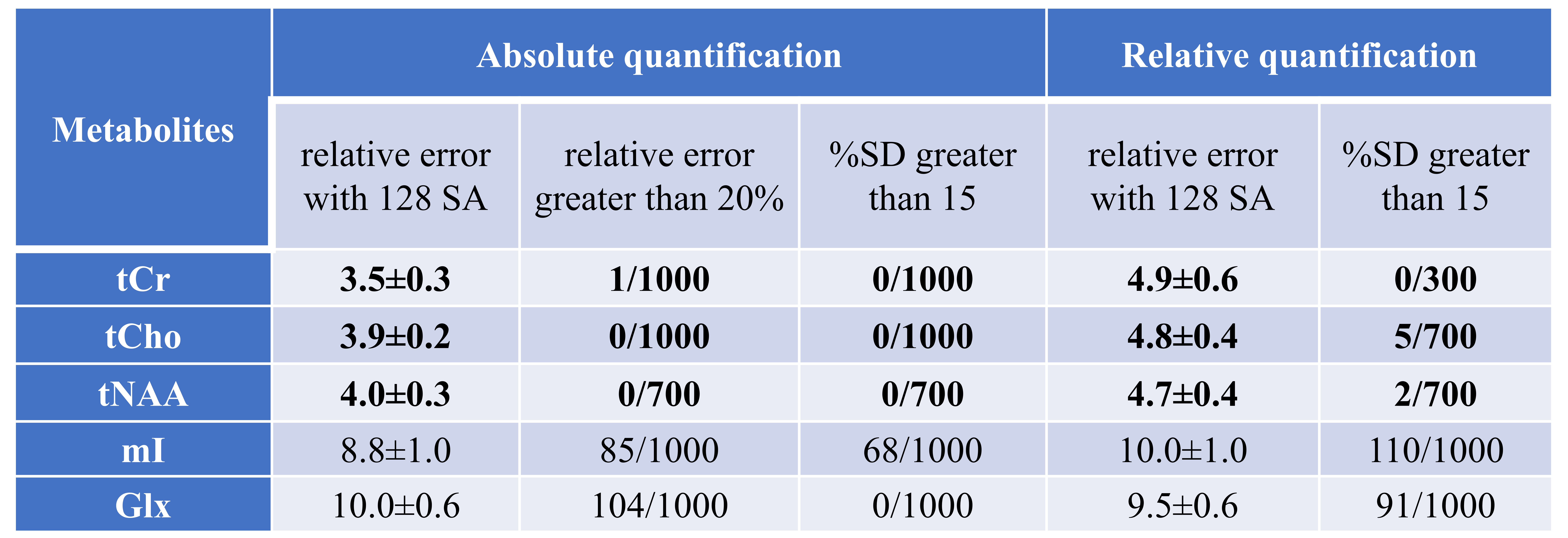
Table 1. Evaluation of ReLSTM denoising results of metabolite concentrations by comparing with the spectrum of 128 NSA (high SNR)
Source
CloudBrian-MRS link: https://csrc.xmu.edu.cn/CloudBrain.html
Acknowledgements
This work was supported in part by National Natural Science Foundation of China (62122064, 61971361, 61871341, 61811530021, 81401405), Natural Science Foundation of Fujian Province of China (2021J011184), Health-Education Joint Research Project of Fujian Province (2019-WJ-31), Nanqiang Outstanding Talent Program of Xiamen University.
References
[1] J.B. Poullet, D.M. Sima, and S. Van Huffel, "MRS signal quantitation: A review of time-and frequency-domain methods," Journal of Magnetic Resonance, vol. 195, no. 2, pp. 134-144, 2008.
[2] S.W. Provencher, "Estimation of metabolite concentrations from localized in vivo proton NMR spectra," Magnetic Resonance in Medicine, vol. 30, no. 6, pp. 672-679, 1993.
[3] J. Near, A.D. Harris, C. Juchem, R. Kreis, M. Marjańska, G. Öz, J. Slotboom, M. Wilson, and C. Gasparovic, "Preprocessing, analysis and quantification in single-voxel magnetic resonance spectroscopy: Experts' consensus recommendations," NMR in Biomedicine, vol. 34, no. 5, pp. 4257, 2020.
[4] C. Choi et al., "Measurement of glycine in the human brain in vivo by 1H‐MRS at 3 T: Application in brain tumors," Magnetic Resonance in Medicine, vol. 66, no. 3, pp. 609-618, 2011.
[5] Q. Yang, Z. Wang, K. Guo, C. Cai and X. Qu, "Physics-Driven Synthetic Data Learning for Biomedical Magnetic Resonance: The imaging physics-based data synthesis paradigm for artificial intelligence," IEEE Signal Processing Magazine, vol. 40, no. 2, pp. 129-140, 2023.
[6] T. Qiu et al., "An automatic denoising method for NMR spectroscopy based on low-rank Hankel model," IEEE Transactions on Instrumentation & Measurement, vol. 70, pp. 1-12, 2021.