物理生成数据学习多场景泛化磁共振成像(中文,English)
王孜1,2, 于小烔1, 王成彦3,4, 陈伟波5, 王家正5, 朱盈樺6, 孙鸿伟7, 李如帅8, 李培勇9, 杨帆10, 韩海伟10, 康泰山11, 林建忠11, 杨晨12, 常书福13, 史张14, 华沙15, 李彦16, 胡娟17, 朱柳红18, 周建军18, 林美金19, 郭杰锋20, 蔡聪波1, 陈忠1, 郭迪21, 杨光 2,22,23,24, 屈小波1,*
1 厦门大学,电子科学系,厦门大学-东软医疗磁共振成像联合研发中心,福建等离子体与磁共振重点研究实验室,健康医疗大数据国家研究院,中国,厦门.
2帝国理工学院, 生物工程系, 英国, 伦敦.
3 上海浦东医院,复旦大学人类表型组研究院, 中国, 上海.
4 国际人类表型组研究院(上海), 中国, 上海.
5 飞利浦医疗科技公司, 中国, 上海.
6 西门子医疗系统有限公司, 中国, 上海.
7 联影智能影像技术研究院, 中国, 北京.
8 南京市第一医院,核医学科, 中国, 南京.
9 山东奥新医疗科技公司,中国, 潍坊.
10 厦门大学附属第一医院, 放射科, 中国, 厦门.
11 厦门大学附属中山医院, 放射科, 中国, 厦门.
12 复旦大学附属中山医院厦门医院, 神经外科, 中国, 厦门.
13 复旦大学附属中山医院, 心血管内科, 中国, 上海.
14 复旦大学附属中山医院, 放射科, 中国, 上海.
15 上海交通大学医学院附属瑞金医院卢湾分院, 心血管内科, 中国, 上海.
16 上海交通大学医学院附属瑞金医院, 放射科, 中国, 上海.
17 昆明医科大学第一附属医院, 医学影像科, 中国, 昆明.
18 复旦大学附属中山医院厦门医院, 放射科, 中国, 厦门.
19 厦门大学, 应用海洋物理与工程系, 中国, 厦门.
20 厦门大学, 微电子与集成电路系, 中国, 厦门.
21 厦门理工学院, 计算机与信息工程学院, 中国, 厦门.
22 帝国理工学院, 国家心肺研究中心, 英国, 伦敦.
23 帝国理工学院, 皇家布朗普顿医院心血管研究中心, 英国, 伦敦.
24 伦敦国王学院,生物医学工程与影像科学学院, 英国, 伦敦.
* Emails: quxiaobo <at> xmu.edu.cn
引用
Zi Wang, Xiaotong Yu, Chengyan Wang, Weibo Chen, Jiazheng Wang, Ying-Hua Chu, Hongwei Sun, Rushuai Li, Peiyong Li, Fan Yang, Haiwei Han, Taishan Kang, Jianzhong Lin, Chen Yang, Shufu Chang, Zhang Shi, Sha Hua, Yan Li, Juan Hu, Liuhong Zhu, Jianjun Zhou, Meijing Lin, Jiefeng Guo, Congbo Cai, Zhong Chen, Di Guo, Guang Yang, Xiaobo Qu, One for multiple: Physics-informed synthetic data boosts generalizable deep learning for fast MRI reconstruction, Medical Image Analysis, 103: 103616, 2025.
概要
磁共振成像(MRI)是重要的无创、无放射性临床诊断的重要工具,为医学诊断提供了丰富多样的人体全身信息,但数据采集时间较长。人工智能(AI)已成为解决快速磁共振成像中重建逆问题的强大工具,但往往专精于特定应用,但尚不能很好地适应复杂的多场景MRI重建的需求。主要原因在于:(1)实测 MRI 数据采集通常耗时、费力且受隐私问题限制。更重要的是,考虑到MRI应用的动态性和多样性,收集涵盖所有成像场景的数据集是不切实际的。(2)训练数据和待重建数据之间的不匹配问题(又称之为分布外数据重建场景)往往是不可避免的,因此AI模型泛化能力的缺乏极大地限制了其在本领域的广泛使用。为了在不同场景中保持良好的重建性能,常见方案是专门针对数据特点训练多个专精AI模型来处理不同任务。如何突破数据瓶颈,实现鲁棒可靠、多场景适用的泛化MRI亟待解决。
主要内容
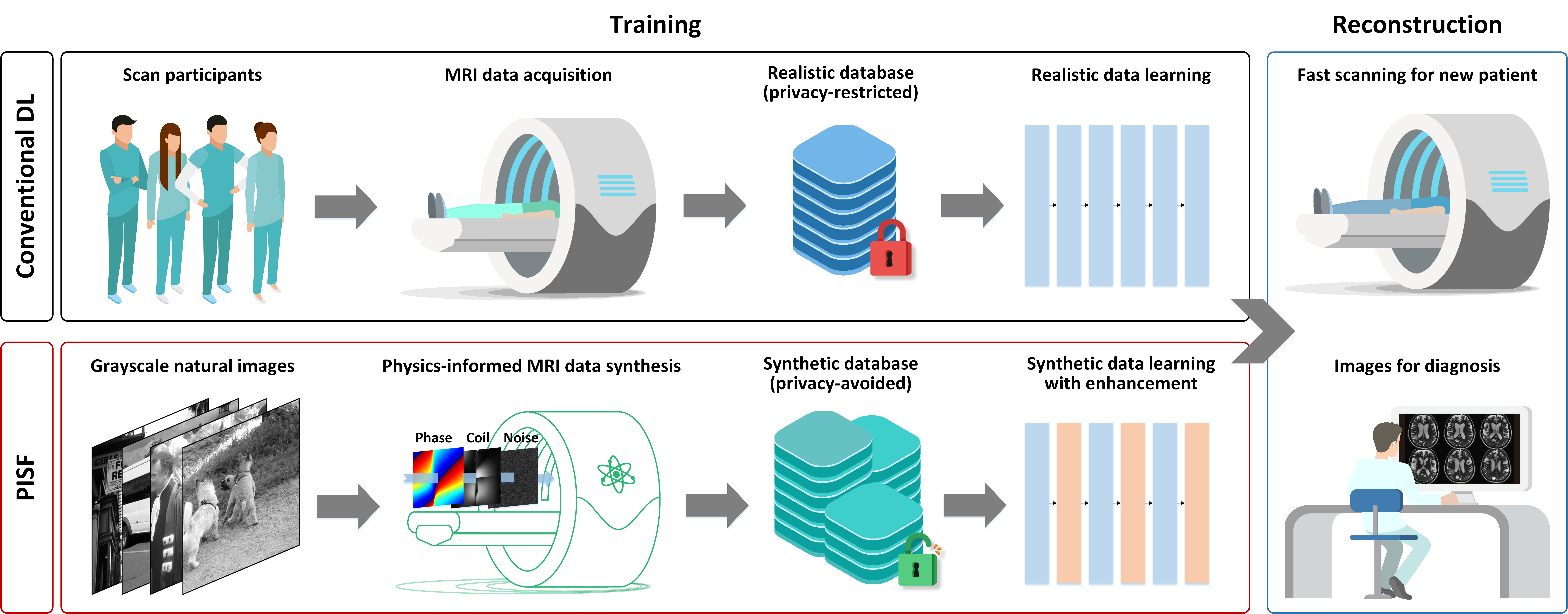
该工作提出了物理启发+生成数据学习+多场景泛化MRI的新范式PISF(图1)。具体来说,基于信号可分离模型和MRI物理生成大规模、多样化的合成数据(图2)训练通用深度图像去伪影模型,显著降低了AI对实测数据的依赖(高达96%)。在重建阶段,通过结合目标数据自适应增强技术进一步校正重建结果,实现了多场景泛化MRI。

图1. 物理生成数据学习助力多场景泛化磁共振成像(PISF)。
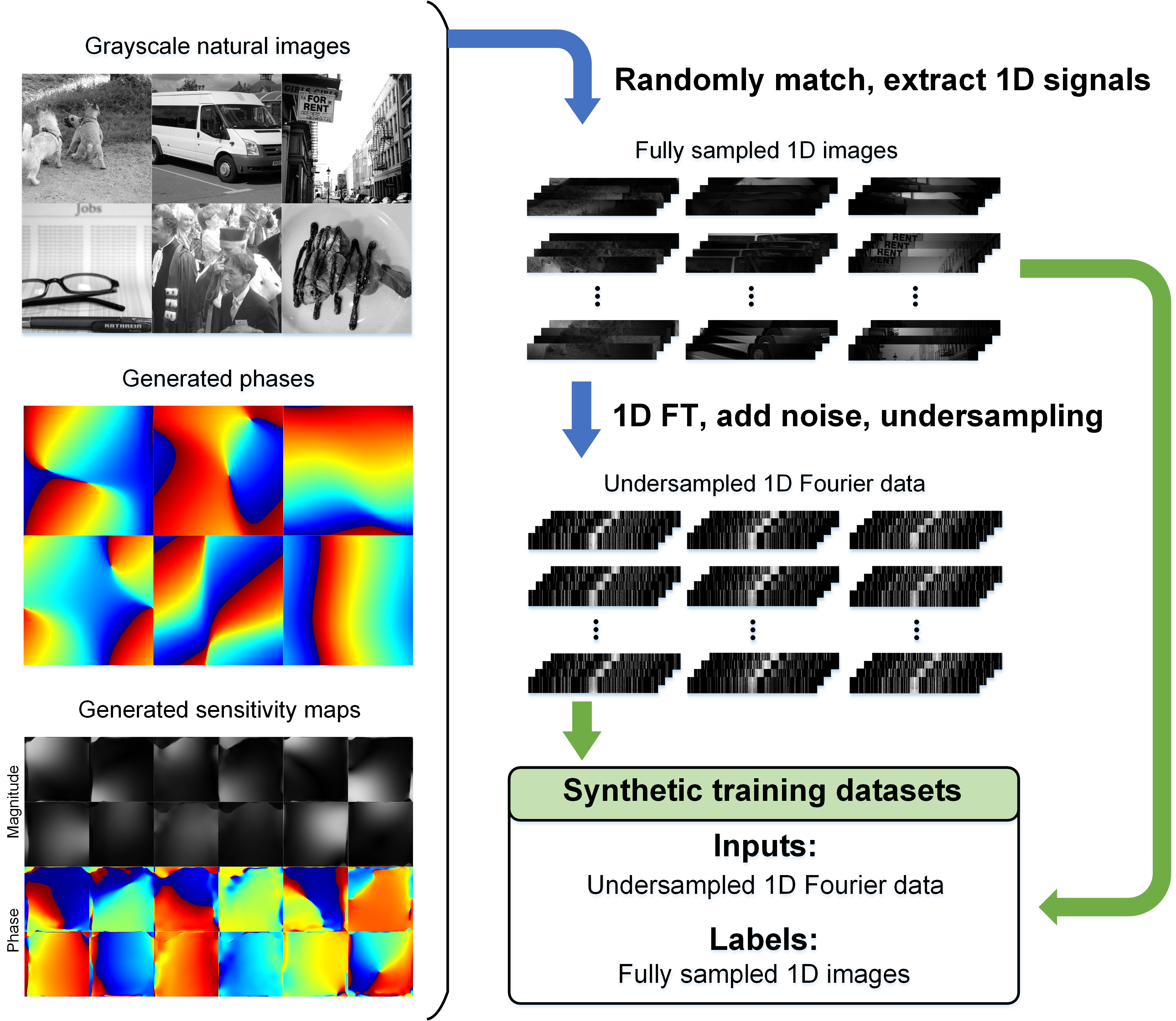
图2. 基于信号可分离模型和MRI物理生成大规模、多样化的合成数据用于AI模型训练。
仅用一个AI模型,所提方法就可以支持5家设备商、7个扫描中心、5种解剖结构、6种图像对比度和4种采样设置的毫秒级高质量泛化重建,并对2种神经疾病和1种心血管疾病的患者数据展现出很好的适应性(图3),10位医生(包括7位放射科医生、1位神经外科医生和2位心内科医生)的图像盲评质量迈入优秀水平(平均为5分制的4.3分)。
此外,还通过对不同类型的训练数据特征进行多角度分析,给出了生成数据泛化学习的全新见解,为AI在MRI中的广泛应用开辟了一条新路径,巧妙规避了传统研究中大规模人体数据采集面临的复杂伦理困境与实际操作难题。
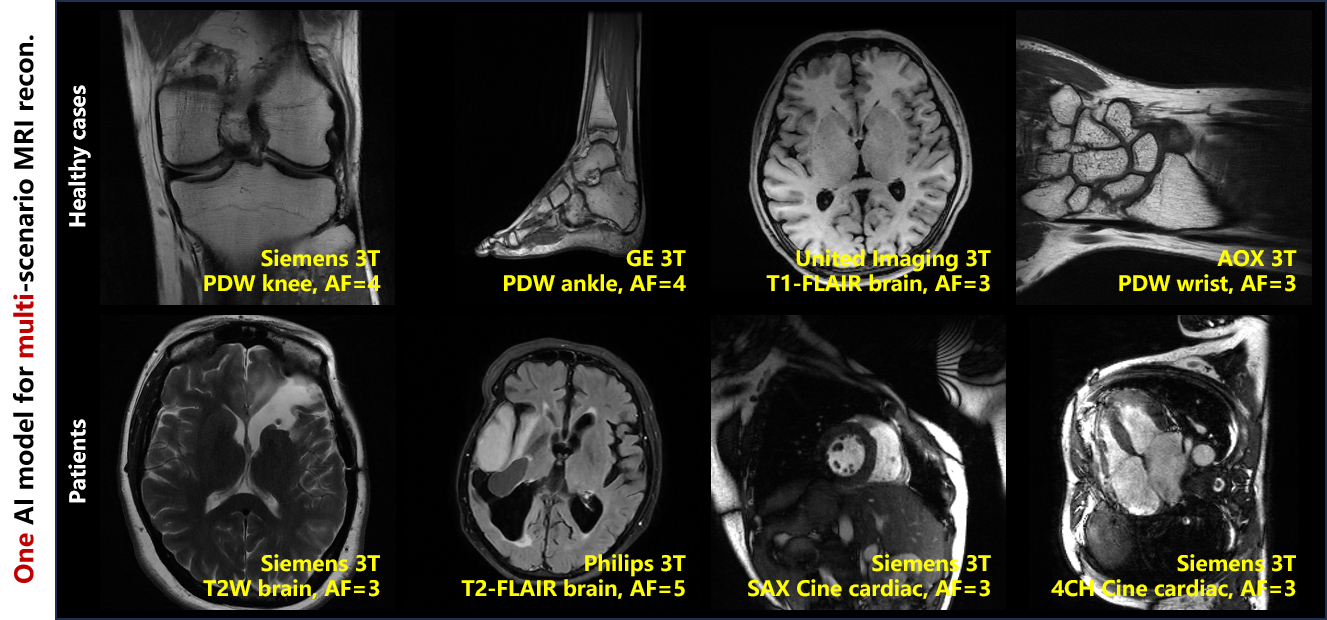
图3. 仅用一个AI模型,可实现多场景MRI的高质量泛化重建。
相关资料
致谢
本工作得到国家自然科学基金(62331021, 62122064, 62371410)、福建省自然科学基金(2023J02005, 2022J011425)、中国教育部产学研合作协同育人项目(231107173160805)、国家重点研发计划(2023YFF0714200)、厦门大学校长基金(20720220063)、英国研究与创新未来领袖基金(MR/V023799/1)、周詠棠高端人才创新团队基金(0621-Z0332004)、厦门大学南强拔尖人才计划、国家留学基金(202306310177)等的资助。
参考文献
[1] P. J. Keall et al., "Integrated MRI-guided radiotherapy — Opportunities and challenges," Nature Reviews Clinical Oncology, vol. 19, no. 7, pp. 458-470, 2022.
[2] M. Lustig, D. Donoho, and J. M. Pauly, "Sparse MRI: The application of compressed sensing for rapid MR imaging," Magnetic Resonance in Medicine, vol. 58, no. 6, pp. 1182-1195, 2007.
[3] Z. Liang, "Spatiotemporal imaging with partially separable functions," in IEEE International Symposium on Biomedical Imaging (ISBI), 2007, pp. 988-991.
[4] M. Lustig and J. M. Pauly, "SPIRiT: Iterative self-consistent parallel imaging reconstruction from arbitrary k-space," Magnetic Resonance in Medicine, vol. 64, no. 2, pp. 457-471, 2010.
[5] J. P. Haldar, "Low-rank modeling of local k-space neighborhoods (LORAKS) for constrained MRI," IEEE Transactions on Medical Imaging, vol. 33, no. 3, pp. 668-681, 2014.
[6] Q. Yang, Z. Wang, K. Guo, C. Cai, and X. Qu, "Physics-driven synthetic data learning for biomedical magnetic resonance: The imaging physics-based data synthesis paradigm for artificial intelligence," IEEE Signal Processing Magazine, vol. 40, no. 2, pp. 129-140, 2023.
[7] H. K. Aggarwal, M. P. Mani, and M. Jacob, "MoDL: Model-based deep learning architecture for inverse problems," IEEE Transactions on Medical Imaging, vol. 38, no. 2, pp. 394-405, 2019.
[8] T. Eo, H. Shin, Y. Jun, T. Kim, and D. Hwang, "Accelerating Cartesian MRI by domain-transform manifold learning in phase-encoding direction," Medical Image Analysis, vol. 63, p. 101689, 2020.
[9] A. Pramanik, H. Aggarwal, and M. Jacob, "Deep generalization of structured low-rank algorithms (Deep-SLR)," IEEE Transactions on Medical Imaging, vol. 39, no. 12, pp. 4186-4197, 2020.
[10] J. Zbontar et al., "fastMRI: An open dataset and benchmarks for accelerated MRI," arXiv: 1811.08839, 2019.
[11] V. Antun, F. Renna, C. Poon, B. Adcock, and A. C. Hansen, "On instabilities of deep learning in image reconstruction and the potential costs of AI," Proceedings of the National Academy of Sciences, vol. 117, no. 48, p. 30088, 2020.
[12] Z. Wang et al., "One-dimensional deep low-rank and sparse network for accelerated MRI," IEEE Transactions on Medical Imaging, vol. 42, no. 1, pp. 79-90, 2023.
[13] G. E. Karniadakis, I. G. Kevrekidis, L. Lu, P. Perdikaris, S. Wang, and L. Yang, "Physics-informed machine learning," Nature Reviews Physics, vol. 3, no. 6, pp. 422-440, 2021.
[14] R. J. Chen, M. Y. Lu, T. Y. Chen, D. F. K. Williamson, and F. Mahmood, "Synthetic data in machine learning for medicine and healthcare," Nature Biomedical Engineering, vol. 5, no. 6, pp. 493-497, 2021.
[15] X. Zhang et al., "A guaranteed convergence analysis for the projected fast iterative soft-thresholding algorithm in parallel MRI," Medical Image Analysis, vol. 69, p. 101987, 2021.
[16] Z. Wang et al., "A sparse model-inspired deep thresholding network for exponential signal reconstruction—Application in fast biological spectroscopy," IEEE Transactions on Neural Networks and Learning Systems, vol. 34, no. 10, pp. 7578-7592, 2023.
[17] T. Zhang, J. M. Pauly, S. S. Vasanawala, and M. Lustig, "Coil compression for accelerated imaging with Cartesian sampling," Magnetic Resonance in Medicine, vol. 69, no. 2, pp. 571-582, 2013.
[18] Y. Zhou et al., "CloudBrain-ReconAI: A cloud computing platform for MRI reconstruction and radiologists' image quality evaluation," IEEE Transactions on Cloud Computing, vol. 12, no. 4, pp. 1359-1371, 2024.
[19] J. P. Haldar and J. Zhuo, "P-LORAKS: Low-rank modeling of local k-space neighborhoods with parallel imaging data," Magnetic Resonance in Medicine, vol. 75, no. 4, pp. 1499-1514, 2016.
[20] A. Sriram et al., "End-to-end variational networks for accelerated MRI reconstruction," in Medical Image Computing and Computer Assisted Intervention (MICCAI), Cham, 2020, pp. 64-73.
[21] O. Jaubert et al., "Training deep learning based dynamic MR image reconstruction using open-source natural videos," Scientific Reports, vol. 14, p. 11774, 2024.
[22] B. Yaman, S. A. H. Hosseini, and M. Akcakaya, "Zero-shot self-supervised learning for MRI reconstruction," in International Conference on Learning Representations (ICLR), 2022, pp. 1-16.
[23] Z. Wang et al., "CMRxRecon2024: A multimodality, multiview k-space dataset boosting universal machine learning for accelerated cardiac MRI," Radiology: Artificial Intelligence, vol. 7, no. 2, p. e240443, 2025.