高保真线圈灵敏度学习高加速磁共振成像(中文,English)
王孜1,#, 方浩铭1,#,钱晨1,施博轩1,包立君1, 朱柳红2,周建军2, 魏文苹3,林建忠4,郭迪5,屈小波1,*
1 厦门大学, 电子科学系, 福建等离子体与磁共振重点研究实验室, 健康医疗大数据国家研究院, 中国, 厦门.
2 复旦大学附属中山医院厦门医院, 放射科, 中国, 厦门.
3 厦门大学附属第一医院, 放射科, 中国, 厦门.
4 厦门大学附属中山医院, 放射科, 中国, 厦门.
5 厦门理工学院, 计算机与信息工程学院, 中国, 厦门.
# 共同第一作者.
* Email: quxiaobo <at> xmu.edu.cn
引用
Zi Wang, Haoming Fang, Chen Qian, Boxuan Shi, Lijun Bao, Liuhong Zhu, Jianjun Zhou, Wenping Wei, Jianzhong Lin, Di Guo, Xiaobo Qu, A Faithful Deep Sensitivity Estimation for Accelerated Magnetic Resonance Imaging , IEEE Journal of Biomedical and Health Informatics, DOI: 10.1109/JBHI.2024.3360128, 2024.
概要
磁共振成像是临床诊断的重要工具,但数据采集时间较长。尽管深度学习在快速成像中以高质量重建图像和重建速度证明了其优越的性能,但大多数方法仍然依赖于预先估计的灵敏度矩阵,线圈灵敏度的估计误差会传导至图像重建环节,严重降低图像的重建质量。因此,可靠的线圈灵敏度估计对于磁共振图像重建至关重要。
主要内容
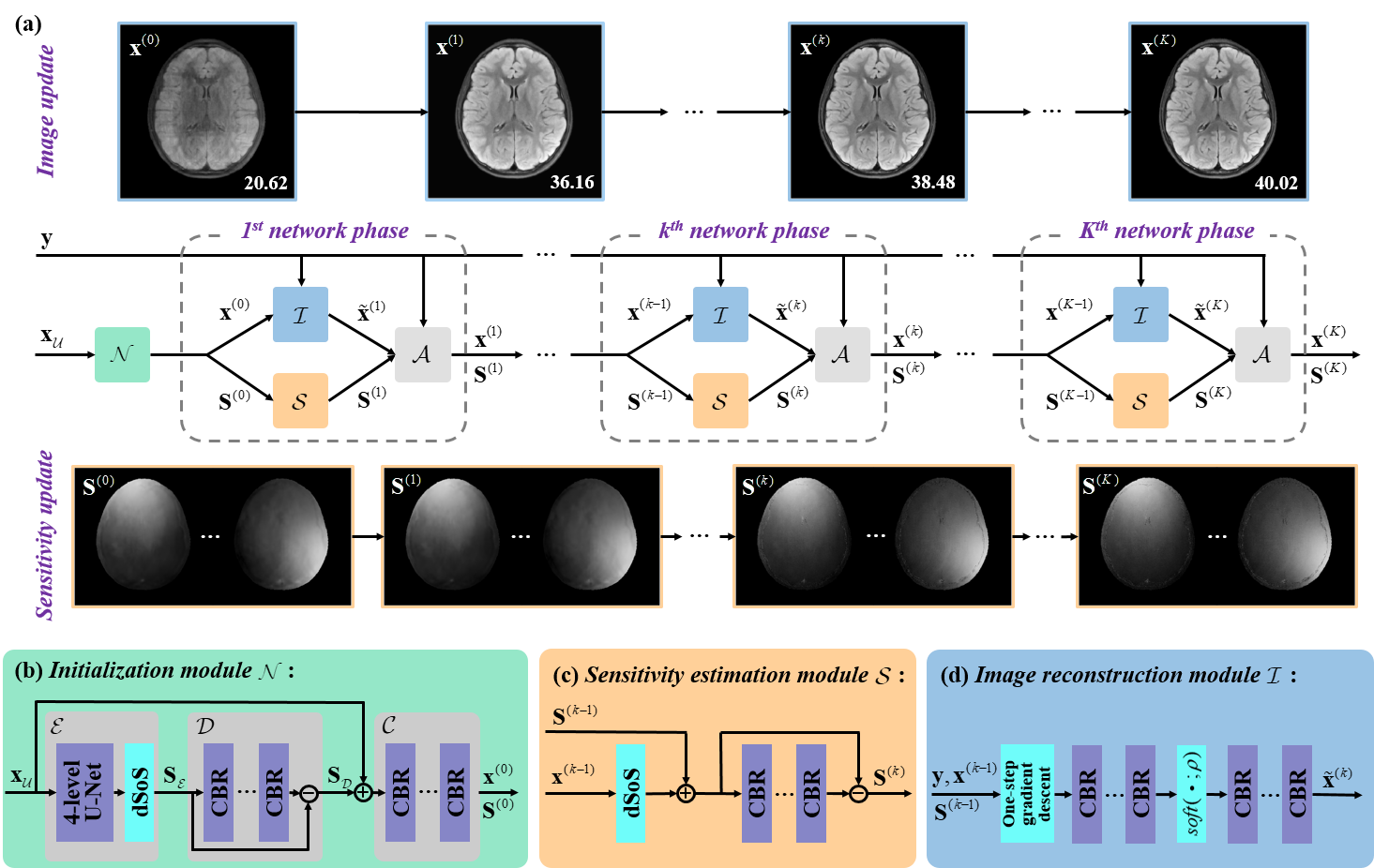
该工作所提的联合线圈灵敏度估计和磁共振图像重建深度学习网络,进一步利用k空间数据提高灵敏度矩阵估计精度并减缓误差在重建环节的传播,通过联合架构在网络中交替迭代校准灵敏度矩阵与更新图像,同时实现了线圈灵敏度的可靠估计和图像的高质量重建(图1)。在实测商用多通道大脑数据上的客观指标和放射科医生的盲评打分表明,在高加速成像(一维欠采6~8倍、二维欠采8~10倍)时,该方法在视觉和定量结果上都实现了比前沿方法更好的图像伪影抑制和细节保留(图2),并对患者数据(病灶更清晰,图3)和校准信号数量(适用无校准重建)具有良好的鲁棒性,3位医生盲评图像质量迈入优秀水平(5分制中的4分,图4)。

图1. 高保真线圈灵敏度学习高加速磁共振成像。
总而言之,该工作提出了一种基于优化模型展开的联合线圈灵敏度估计与图像重建的深度学习方法。该方法在重建过程中,逐步提供高保真的具有高频信息的灵敏度矩阵,以进一步校正重建图像,实现了高加速倍数下的毫秒级临床适用成像,患者数据适应性好,医生盲评图像质量佳。此外,还通过中间结果可视化以理解网络行为,清晰揭示了灵敏度矩阵和重建图像相互校正的过程。
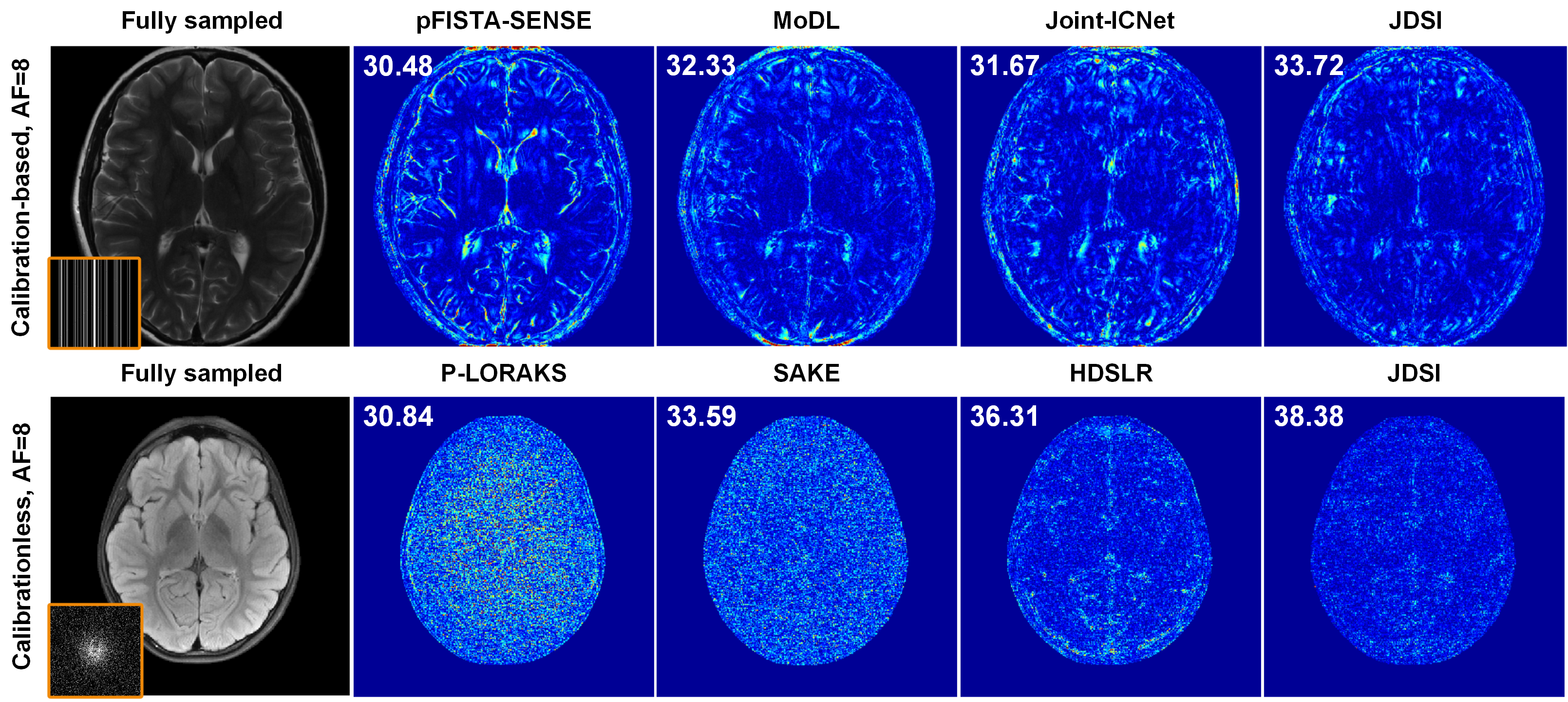
图2. 高加速成像下的大脑重建结果,包含有校准重建和无校准重建。重建图左上角为峰值信噪比PSNR(dB)。
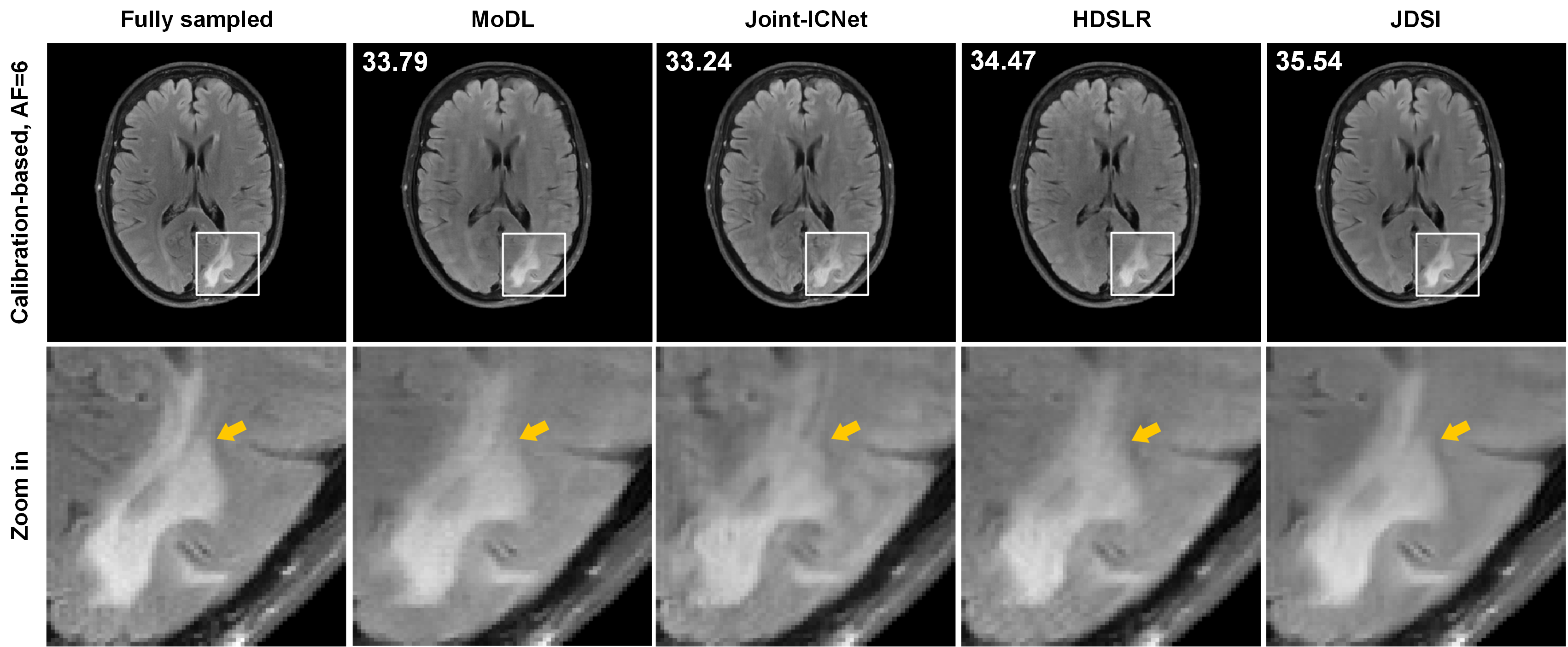
图3. 大脑白质损伤患者数据重建结果。黄色箭头标记的是病灶。重建图左上角为峰值信噪比PSNR(dB)。
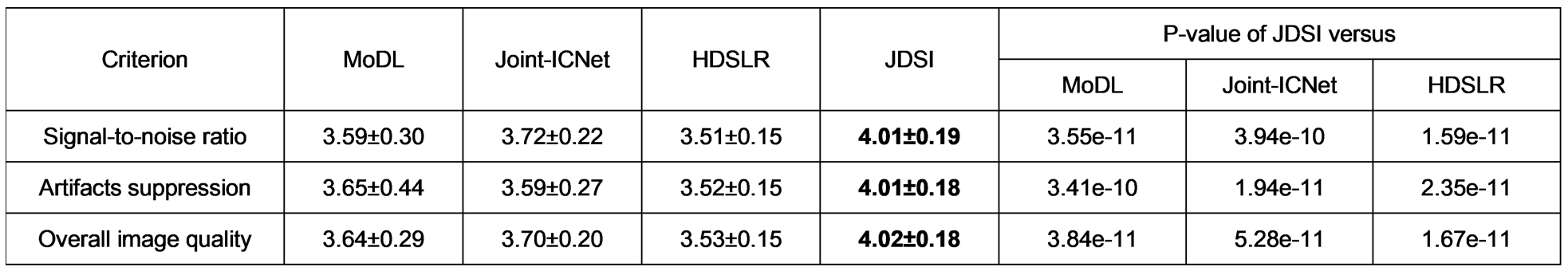
图4. 三位医生图像质量盲评打分结果。采用5分制,分数越高代表对应指标越好。P值小于0.01代表两个对比方法间具有显著统计学差异。
相关资料
致谢
本工作得到国家自然科学基金(62122064, 62331021, 62371410和62071405)、福建省自然科学基金(2023J02005和2021J011184)、厦门大学校长基金(20720220063)、厦门大学南强拔尖人才计划、国家留学基金(202306310177)的资助。
参考文献
[1] G. Harisinghani Mukesh, A. O’Shea, and R. Weissleder, "Advances in clinical MRI technology," Science Translational Medicine, vol. 11, no. 523, p. eaba2591, 2019.
[2] M. Lustig, D. Donoho, and J. M. Pauly, "Sparse MRI: The application of compressed sensing for rapid MR imaging," Magnetic Resonance in Medicine, vol. 58, no. 6, pp. 1182-1195, 2007.
[3] S. Wang et al., "Accelerating magnetic resonance imaging via deep learning," in IEEE 13th International Symposium on Biomedical Imaging (ISBI), 2016, pp. 514-517.
[4] B. Zhu, J. Z. Liu, S. F. Cauley, B. R. Rosen, and M. S. Rosen, "Image reconstruction by domain-transform manifold learning," Nature, vol. 555, no. 7697, pp. 487-492, 2018.
[5] Z. Wang et al., "One-dimensional deep low-rank and sparse network for accelerated MRI," IEEE Transactions on Medical Imaging, vol. 42, no. 1, pp. 79-90, 2023.
[6] Q. Yang, Z. Wang, K. Guo, C. Cai, and X. Qu, "Physics-driven synthetic data learning for biomedical magnetic resonance: The imaging physics-based data synthesis paradigm for artificial intelligence," IEEE Signal Processing Magazine, vol. 40, no. 2, pp. 129-140, 2023.
[7] L. Ying and J. Sheng, "Joint image reconstruction and sensitivity estimation in SENSE (JSENSE)," Magnetic Resonance in Medicine, vol. 57, no. 6, pp. 1196-1202, 2007.
[8] F. K. Zbontar et al., "FastMRI: An open dataset and benchmarks for accelerated MRI," arXiv: 1811.08839, 2019.
[9] X. Zhang et al., "A guaranteed convergence analysis for the projected fast iterative soft-thresholding algorithm in parallel MRI," Medical Image Analysis, vol. 69, 101987, 2021.
[10] H. K. Aggarwal, M. P. Mani, and M. Jacob, "MoDL: Model-based deep learning architecture for inverse problems," IEEE Transactions on Medical Imaging, vol. 38, no. 2, pp. 394-405, 2019.
[11] Y. Jun, H. Shin, T. Eo, and D. Hwang, "Joint deep model-based MR image and coil sensitivity reconstruction network (Joint-ICNet) for fast MRI," in IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2021, pp. 5270-5279.
[12] W. Zhou, A. C. Bovik, H. R. Sheikh, and E. P. Simoncelli, "Image quality assessment: from error visibility to structural similarity," IEEE Transactions on Image Processing, vol. 13, no. 4, pp. 600-612, 2004.
[13] Y. Zhou et al., "XCloud-pFISTA: A medical intelligence cloud for accelerated MRI," in 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 2021, pp. 3289-3292.
[14] M. Uecker et al., "ESPIRiT—An eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA," Magnetic Resonance in Medicine, vol. 71, no. 3, pp. 990-1001, 2014.
[15] A. Sriram et al., "End-to-End variational networks for accelerated MRI reconstruction," in Medical Image Computing and Computer Assisted Intervention (MICCAI), 2020, pp. 64-73.