An Automatic Denoising Method for NMR Spectroscopy Based on Low-Rank Hankel Model (中文,English)
Tianyu Qiu1, Wenjing Liao2, Yihui Huang1, Jinyu Wu1, Di Guo3, Dongbao Liu1, Xin Wang1, Jian-Feng Cai4, Bingwen Hu5, Xiaobo Qu1,*
1 Fujian Provincial Key Laboratory of Plasma and Magnetic Resonance, Biomedical Intelligent Cloud Research and Development Center, Department of Electronic Science, Xiamen University, Xiamen 361005, China.
2 School of Mathematics, Georgia Institute of Technology, Atlanta, GA 30332 USA.
3 School of Computer and Information Engineering, Fujian Provincial University Key Laboratory of Internet of Things Application Technology, Xiamen University of Technology, Xiamen 361024, China.
4 Department of Mathematics, The Hong Kong University of Science and Technology, Hong Kong, China.
5 Department of Physics, East China Normal University, Shanghai 200062, China.
* Emails: quxiaobo <at> xmu.edu.cn or quxiaobo2009 <at> gmail.com
Citation
Tianyu Qiu, Wenjing Liao, Yihui Huang, Jinyu Wu, Di Guo, Dongbao Liu, Xin Wang, Jian-Feng Cai, Bingwen Hu, Xiaobo Qu, An Automatic Denoising Method for NMR Spectroscopy Based on Low-Rank Hankel Model, IEEE Transactions on Instrumentation & Measurement, 70: 1-12, 2021.
Synopsis
In nuclear magnetic resonance (NMR) spectroscopy, denoising is an important task due to its poor sensitivity. In this work, noise removal serves as a low rank Hankel matrix reconstruction from noisy free induction decay (FID) signals, since noiseless FID signals are commonly modeled as exponential functions and their Hankel matrices are usually low rank. A faithful denoising mainly depends on the regularization parameter helping to distinguish meaningful signal from noise. We further derived a theoretical bound to automatically set this parameter. Numerical experiments on the realistic NMR spectroscopy data show that noise can be effectively removed with the proposed approach.
Method
1. Background
Nuclear magnetic resonance (NMR) spectroscopy has grown into an essential tool for biomedical studies [1-3]. Unfortunately, NMR spectroscopy are usually corrupted by Gaussian noise in practice. One of common approaches to denoise is to average multiple signal acquisitions. However, the multiple acquisitions are not always available or too costly in real applications.
The low-rank Hankel property of exponential signals plays an important role in the denoising of NMR spectroscopy, such as Low-Rank Hankel Matrix reconstruction method (LRHM) [4]. Nevertheless, selecting a proper regularization parameter is often based on personal experience.
Therefore, we propose a denoising method, named as CHORD, with an auto-setting parameter based on LRHM. An accurate estimate on the spectral norm of weighted Hankel matrices is provided as a guidance to set the regularization parameter. The bound can be efficiently calculated since it only depends on the standard deviation of the noise and a constant. Aided by the bound, one can easily obtain an auto-setting regularization parameter to produce promising denoised results.
2. Convex Hankel low-rank matrix approximation for denoising exponential signals
In NMR spectroscopy, FID signals are commonly modeled as a sum of R exponentials with decay [4][5][10-14]
$$
x_{0}(t_{n})=\sum_{r=1}^{R}a_{r}e^{\left({j2{\pi}f_{r}-{\tau}_{r}}\right)t_{n}}, \ n=0,\ldots,2N, (1)
$$
where $ a_{r} $ denotes the signal amplitude, $ f_{r} $ is the central frequency, and $ {\tau}_{r} $ is the decay factor.
The proposed denoising method calls Convex Hankel lOw-Rank matrix approximation for Denoising exponential signals (CHORD), where one solves the following optimization problem:
$$
\mathbf{\hat{x}}=\arg\min_{\mathbf{x} \in \mathbb{C}^{2N+1}} \left\|{\mathcal{R}\mathbf{x}}\right\|_{*}+\frac{\lambda}{2}\left\|{\mathbf{y}-\mathbf{x}}\right\|_{2}^{2}, (2)
$$
where $\mathbf{y}{\in}\mathbb{C}^{(2N+1){\times}1}$ denotes the observed signal contaminated by Gaussian noise $\mathbf{z}$. The operator $\mathcal{R}$ transforms a vector into a Hankel matrix. λ denotes the regularization parameter.
According to the subgradient of the nuclear norm [6] and some approximation, the proper λ is chosen as
$$
\lambda \leq {\frac{1}{\left(\left|\left\|\mathbf{Z}\right\|_{2}-\left\|\mathbf{\tilde{X}}\right\|_{2}\right|\right)}}, (3)
$$
where $\mathbf{Z}$ denotes a weighted Hankel matrix converted by $\mathbf{z}$
$$
\mathbf{Z}=
\left(
\begin{array}{cccc}
z_1 & \frac{z_2}{2} & \cdots & \frac{z_{N+1}}{N+1} \\
\frac{z_2}{2} & \frac{z_3}{3} & \cdots & \frac{z_{N+2}}{N} \\
\vdots & \vdots & \cdots & \vdots \\
\frac{z_{N+1}}{N+1} & \frac{z_{N+2}}{N} & \cdots & z_{2N+1}
\end{array}
\right), (4)
$$
and similarly, $\mathbf{\tilde{X}}$ denotes a weighted Hankel matrix converted by $\mathbf{x}_{0}-\mathbf{\hat{x}}$, where $\mathbf{\hat{x}}$ is the minimizer of (2).
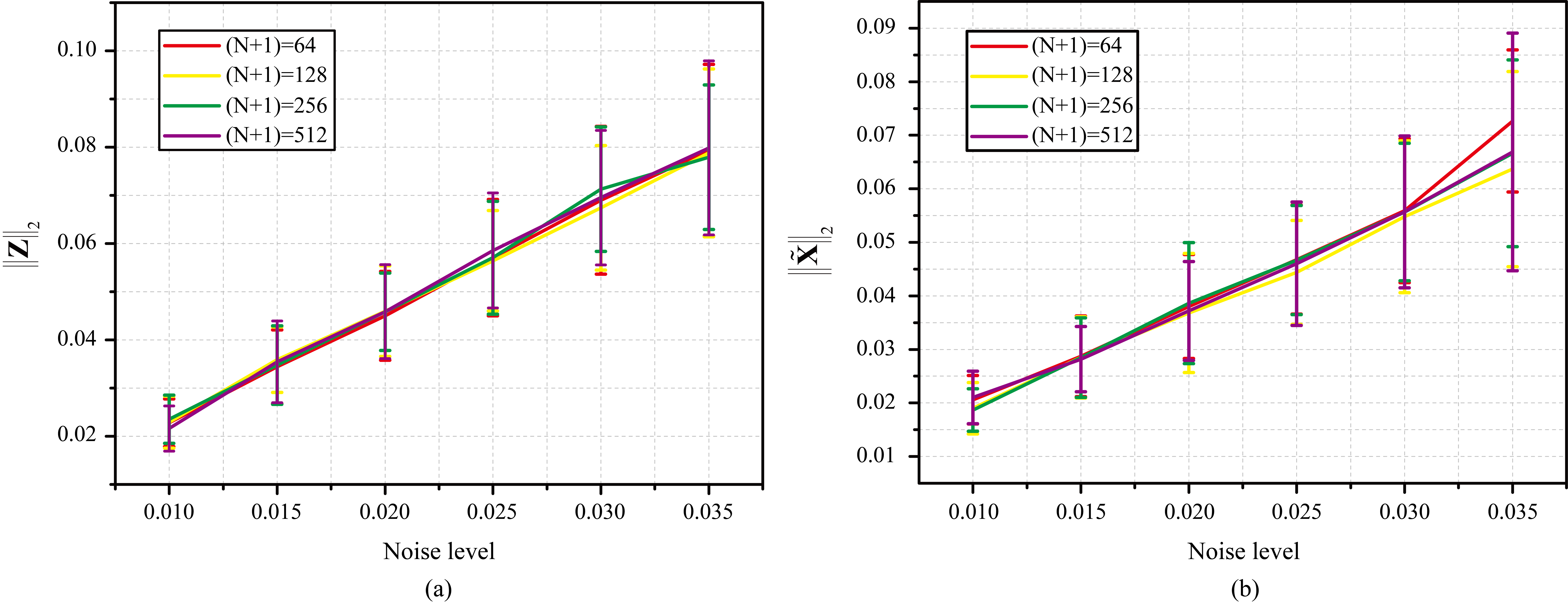
By observing the relationship among the spectral norm of weighted Hankel matrices, the noise level, and the size of the matrix, we find that the empirical means of $\left\|{\mathbf{Z}}\right\|_2$ and $\left\|{\mathbf{\tilde{X}}}\right\|_2$ are almost independent of N (See Fig. 1). Furthermore, these empirical means increase as the increasing of the standard deviation σ of the noise.
In applications, it is expected that signal details can be preserved as much as possible, thus we propose to select the regularization parameter as
$$
{\lambda}^{*}=\frac {1} {\left|\mathbb{E}\left\|\mathbf{Z}\right\|_2-\mathbb{E}\left\|\mathbf{\tilde{X}}\right\|_2\right|}. (5)
$$

Fig. 1. Relation between the spectral norm and the standard deviation σ of the Gaussian noise $\mathbf{z}$. (a) Relation between $\left\|{\mathbf{Z}}\right\|_2$ and σ in 100 Monte Carlo trials. (b) Relation between $\left\|{\mathbf{\tilde{X}}}\right\|_2$ and σ in 50 Monte Carlo trials. Matrices are of size (N+1)×(N+1) with (N+1)=64, 128, 256, and 512, respectively. The curve represents the mean of the spectral norm versus σ, and the standard deviation of the spectral norm is indicated by the vertical bar. Note: $\mathbf{x}_0$ are damped exponential signals with random $a_r$, $f_r$, and $\tau_r$. $\mathbf{\hat{x}}$ is obtained from CHORD. The parameter λ is chosen such that the NRMSE is minimized.
We prove that the bound estimate of $\mathbb{E}\left\|{\mathbf{Z}}\right\|_2$ is as follows
$$
{\sigma}\frac{C(N+1)}{2N+1}\sqrt{R_{N}^{2}\left({1+\log\frac{R_{N}^{4}}{Q_{N}^{4}}}\right)}{\leq}\mathbb{E}\left\|{\mathbf{Z}}\right\|_{2}{\leq}{\sigma}\sqrt{2C_{\mathbf{w}}\log\left({2N+2}\right)}, (6)
$$
where $C_{\mathbf{w}} = \max(\sum_{k=0}^{N}{w_{k}^{-2}},\sum_{k=1}^{N+1}{w_{k}^{-2}},\ldots,\sum_{k=N}^{2N}{w_{k}^{-2}})$ with the weight $\mathbf{w}\in\mathbb{R}^{(2N+1)\times{1}}$ defined as
$\mathbf{w}=\left[{
\begin{array}{ccccccc}
1 & 2 & \cdots & N+1 & \cdots & 2 & 1
\end{array}
}\right]^{T}$. $R_{N}$ and $Q_{N}$ are sequences defined as $R_{N}^{2}=\sum_{k=0}^{2N}\left|{d_k}\right|^{2}$ and $Q_{N}^{4}=\sum_{k=0}^{2N}\left|{d_k}\right|^{4}$, where$ d_{k}=
\left\{
\begin{array}{ll}
\frac{2}{(k+1)(k+2)}\sum_{m=0}^{k}\frac{1}{m+1}, 0\leq k\leq N \\
\frac{2}{(2N-k+1)(k+2)}\sum_{m=k}^{2N}\frac{1}{m-N+1}, N< k\leq 2N \\
\end{array}
\right. $. When $N$ is large enough, the upper bound and the lower bound only differ by a factor of $\sqrt{\log N}$.
Choosing the lower bound of $\mathbb{E}\left\|\mathbf{Z}\right\|_{2}$ tends to obtain a relatively large $\lambda$, which is beneficial to preserve more signal details. Therefore, we choose the lower bound. Regarding the constant $C$, we suggest that $C=2.9$ for denoising.
We perform experiments with different $N$, σ, λ, signals, and noises in order to determine a proper empirical estimate value of $\left\|\mathbf{\tilde{X}}\right\|_2$. Results of Monte Carlo trials indicate that σ is the main factor that determines $\mathbb{E}\left\|\mathbf{\tilde{X}}\right\|_2$. Moreover, this empirical mean of $\mathbf{\tilde{X}}$ seems to be linear to σ. We suggest $\mathbb{E}\left\|\mathbf{\tilde{X}}\right\|_2=1.94σ$ for denoising.
3. Main result
Here we adopt CHORD to denoise a 1D NMR spectrum, and compare the denoised results with the typical method, Cadzow [7][8], and the state-of-the-art method, rQRd [7].
The real data is a 1D 1H NMR spectrum that was acquired at 298 K on a Varian 500 MHz (11.7 T) magnetic resonance system (Agilent Technologies, Santa Clara, CA, USA) equipped with a 5-mm indirect detection probe. A 8.3 μs single-pulse sequence was used, and 64 scans were acquired. The total acquisition time took 286.7s. The concentration of creatine, choline, magnesium citrate, and calcium citrate are 0.03, 0.03, 0.06, and 0.06 g/mL, respectively.
We truncate the signal from the end of noisy FID to estimate the standard deviation of the noise to mimic the real cases. The truncated length is verified by Kolmogorov–Smirnov (KS) test [9].
The denoised spectra are presented in Fig. 2. Under a relatively strong noise level (σ = 0.035), Cadzow smooths the spectrum, which, on the one side, offers a nice denoised results, on the other side, however, leads to the missing of some peaks (such as the peaks at 6.8 ppm). rQRd provides a spectrum with obvious noise [orange lines] and weakens low-intensity peaks (such as the peaks at 6.8 ppm). CHORD is capable of effectively removing noise and keeping more details of peaks.
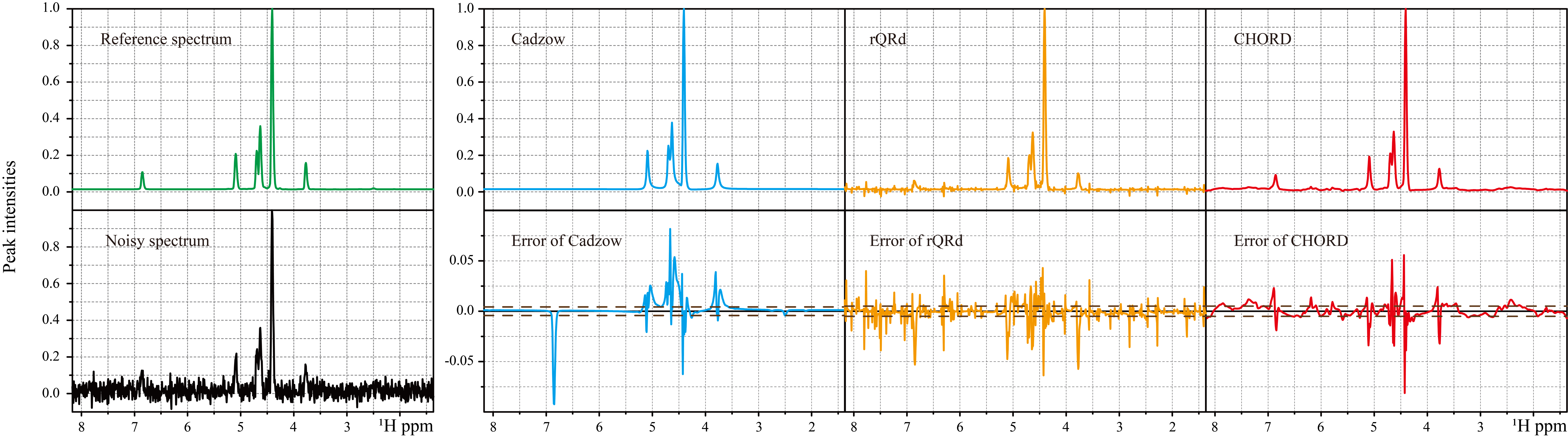
Denoised results of realistic metabolite spectrum with σ = 0.035. The green lines denote the reference spectrum with high SNR. The black lines indicate noisy spectra. The blue, orange, and red lines are denoised results of Cadzow, rQRd, and CHORD, respectively. Note: The results of Cadzow and rQRd that enable the lowest NRMSE are presented here. The brown long dash lines denote the MAEs of the denoised spectra.
Download
The MATLAB code of CHORD can be downloaded here.
This work also can be downloaded here.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China under Grant 6212200447, Grant 61971361, Grant 61871341, and Grant 61811530021; in part by the National Key Research and Development Program of China under Grant 2017YFC0108703; in part by the Science and Technology Program of Xiamen under Grant 3502Z20183053; in part by the Health-Education Joint Research Project of Fujian Province under Grant 2019-WJ-31; and in part by Xiamen University Nanqiang Outstanding Talents Program.
References:
[1] A. Wegemann, et al., "A portable NMR spectrometer with a probe head combining RF and DC capabilities to generate pulsed-field gradients," IEEE Trans. Instrum. Meas., vol. 69, no. 10, pp. 8628–8636, Oct. 2020.
[2] O. Beckonert, et al., "High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues," Nature Protocols, vol. 5, no. 6, pp. 1019–1032, Jun. 2010.
[3] M. C. Preul, et al., "Accurate, noninvasive diagnosis of human brain tumors by using proton magnetic resonance spectroscopy," Nature Med., vol. 2, no. 3, pp. 323–325, Mar. 1996.
[4] X. Qu, et al., "Accelerated NMR spectroscopy with low-rank reconstruction," Angew. Chem. Int. Edit., vol. 54, no. 3, pp. 852–854, Jan. 2015.
[5] T. Qiu, et al., "Review and prospect: NMR spectroscopy denoising and reconstruction with low-rank Hankel matrices and tensors," Magn. Reson. Chem., vol. 59, pp. 324–345, Mar. 2021.
[6] J.-F. Cai, et al., "A singular value thresholding algorithm for matrix completion," SIAM J. Optim., vol. 20, no. 4, pp. 1956–1982, 2010.
[7] L. Chiron, et al., "Efficient denoising algorithms for large experimental datasets and their applications in Fourier transform ion cyclotron resonance mass spectrometry," Proc. Nat. Acad. Sci. USA, vol. 111, no. 4, pp. 1385–1390, Jan. 2014.
[8] J. Gillard, "Cadzow’s basic algorithm, alternating projections and singular spectrum analysis," Statist. Interface, vol. 3, no. 3, pp. 335–343, 2010.
[9] F. J. Massey Jr., "The Kolmogorov-Smirnov test for goodness of fit," J. Amer. Statist. Assoc., vol. 46, no. 253, pp. 68–78, 1951.
[10] J. Ying, et al., "Hankel matrix nuclear norm regularized tensor completion for N-dimensional exponential signals," IEEE Trans. Signal Proces., vol. 65, no. 14, pp. 5520-5533, 2017.
[11] J. Ying, et al., "Vandermonde factorization of Hankel matrix for complex exponential signal recovery—Application in fast NMR spectroscopy," IEEE Trans. Signal Proces., vol. 66, no. 21, pp. 5520–5533, 2018.
[12] H. Lu, et al., "Low rank enhanced matrix recovery of hybrid time and frequency data in fast magnetic resonance spectroscopy," IEEE Trans. Biomed. Eng., vol. 65, no. 4, pp. 809–820, 2017.
[13] D. Guo, et al., "A fast low rank Hankel matrix factorization reconstruction method for non-uniformly sampled magnetic resonance spectroscopy," IEEE Access, vol. 5, pp. 16033-16039, 2017.
[14] D. Chen, et. al., "Review and prospect: Deep learning in nuclear magnetic resonance spectroscopy," Chem. Eur. J., vol. 26, no. 46, p. 10, Aug. 2020.