综述与展望:深度学习核磁共振波谱
陈棣成1,#, 王孜1,#, 郭迪2, Vladislav Orekhov3, 屈小波1,*
1厦门大学,电子科学系,福建等离子体与磁共振重点研究实验室,中国,厦门,361005
2厦门理工学院,计算机与信息工程学院,中国,厦门,361024
3哥德堡大学,化学与分子生物学学院,瑞典,哥德堡,40530
*邮箱:quxiaobo <at> xmu.edu.cn or quxiaobo2009 <at> gmail.com
#共同第一作者
引用
Dicheng Chen#, Zi Wang#, Di Guo, Vladislav Orekhov, Xiaobo Qu*. Review and Prospect: Deep Learning in Nuclear Magnetic Resonance Spectroscopy, Chemistry -A European Journal, DOI: 10.1002/chem.202000246, 2020.
链接
http://dx.doi.org/10.1002/chem.202000246
概要
核磁共振波谱能够提供分子结构的原子水平的信息,是分析分子动力学和相互作用不可缺少的工具,在化学、生命科学等领域应用十分广泛。深度学习提供了一种前所未有的分析和处理数据的方法,鉴于它的成功,核磁共振领域的研究人员开始关注深度学习技术,并尝试用它来解决传统方法的不足之处。
该论文首次系统地综述了深度学习在核磁共振波谱重建、去噪、化学位移预测和自动选峰等方面的应用,并展望了深度学习作为该领域的一种全新方法的前景,以期望将核磁共振波谱转化为化学和生命科学中更有效、更强大的工具。
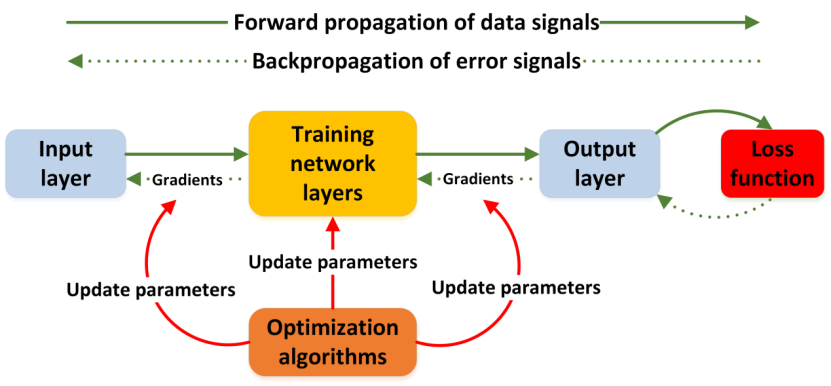
图1 深度学习神经网络训练流程图
主要内容
首先,深度学习已被证明可以非均匀欠采样数据中快速重建高质量波谱。该论文分别评述了基于卷积神经网络和长短时记忆网络的两种深度学习方法,同时两种方法都只需使用仿真的指数信号进行网络训练。结果表明,其波谱重建的性能不仅可以与目前最前沿的迭代方法相媲美,还在重建时间上具有突出的优势。
其次,深度学习已被有效应用于核磁共振波谱的去噪。该论文介绍了一种基于卷积神经网络的方法,它只需使用仿真的脑部谱信号进行网络训练,就可以得到具有大量干扰的频谱与高信噪比频谱之间的映射关系。鉴于其对低信噪比的鲁棒性,有望将促进临床应用的发展。
第三,该论文总结了多种用来建立化合物信息与其化学位移之间的关系的神经网络方法。结果表明,这些方法在化学位移预测方面明显接近经验方法的极限,同时其精度可与量子化学计算方法相媲美。
最后,深度学习还可用于自动选峰,展现了其在自动化复杂数据分析方面的巨大潜力。该论文主要关注基于卷积神经网络和全连接网络的两种深度学习方法。研究结果显示,自动谱峰选取的精度很高,与通过专家人工选取的信号区域基本一致。
此外,针对核磁共振波谱领域的现状,该论文提出了深度学习未来面临的挑战:模型可解释性不强、训练数据集不充足等,并展望了未来可解决的问题:高维波谱重建、新化合物结构预测、功能性波谱应用等。
致谢
这项工作得到了国家自然科学基金(61971361、61871341、U1632274),国家自然科学基金国际合作与交流项目(61811530021),国家重点研发计划(2017YFC0108703),福建省自然科学基金(2018J06018),中央高校基本科研基金(20720180056),厦门大学南强拔尖人才计划,厦门市科学技术计划(3502Z20183053)、瑞典研究委员会基金(2015–04614)和瑞典战略研究基金(ITM17-0218)的资助。
参考文献
[1] a) B. R. Kowalski, C. F. Bender, Anal. Chem. 1972, 44, 1405-1411; b) K. Esbensen, P. Geladi, J. Chemometr. 1990, 4, 389-412.
[2] Y. LeCun, Y. Bengio, G. Hinton, Nature 2015, 521, 436.
[3] a) S. Wang, Z. Su, L. Ying, X. Peng, S. Zhu, F. Liang, D. Feng, D. Liang, in 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), 2016, pp. 514-517; b) C. Cai, C. Wang, Y. Zeng, S. Cai, D. Liang, Y. Wu, Z. Chen, X. Ding, J. Zhong, Magn. Reson. Med. 2018, 80, 2202-2214; c) J. Schlemper, J. Caballero, J. V. Hajnal, A. N. Price, D. Rueckert, IEEE Trans. Med. Imaging 2018, 37, 491-503; d) Y. Yang, J. Sun, L. I. H, Z. Xu, IEEE Trans. Pattern Anal. and Mach. Intell. 2020, 42, 521-538.
[4] a) X. Qu, Y. Huang, H. Lu, T. Qiu, D. Guo, T. Agback, V. Orekhov, Z. Chen, Angew. Chem. Int. Ed. 2019, DOI: 10.1002/anie.201908162; b) D. F. Hansen, J. Biomol. NMR 2019, 73, 577-585.
[5] P. Klukowski, M. Augoff, M. Zięba, M. Drwal, A. Gonczarek, M. J. Walczak, Bioinformatics 2018, 34, 2590-2597.
[6] D. Svozil, V. Kvasnicka, J. í. Pospichal, Chemometr. Intell. Lab. 1997, 39, 43-62.
[7] a) Y. Lecun, B. Boser, J. Denker, D. Henderson, R. E. Howard, W. Hubbard, L. Jackel, in Advances in Neural Information Processing Systems (NIPS), 1990; b) S. Lawrence, C. Giles, A. Tsoi, A. Back, IEEE Trans. Neural Networks 1997, 8, 98-113; c) A. Krizhevsky, I. Sutskever, G. Hinton, in Neural Information Processing Systems (NIPS) 2012.
[8] a) Y. Bengio, P. Simard, P. Frasconi, IEEE Trans. Neural Networks 1994, 5, 157-166; b) R. Williams, D. Zipser, Neural Comput. 1989, 1, 270-280; c) S. Hochreiter, J. Schmidhuber, Neural Comput. 1997, 9, 1735-1780.
[9] N. Hecht, in International 1989 Joint Conference on Neural Networks, 1989, pp. 593-605.
[10] L. Bottou, P. Neuro-Nımes, 1991, 91.
[11] D. Kingma, J. Ba, Computer Science 2014.
[12] N. Srivastava, G. Hinton, A. Krizhevsky, I. Sutskever, R. Salakhutdinov, J. Mach. Learn. Res. 2014, 15, 1929-1958.
[13] S. Ioffe, C. Szegedy, Computer Science 2015.
[14] V. Nair, G. Hinton, Proceedings of the 27th International Conference on Machine Learning (ICML10), 2010, 27, 807–814.
[15] S. Min, B. Lee, S. Yoon, Brief. Bioinform. 2016, 18, 851-869.
[16] M. Abadi, A. Agarwal, P. Barham, E. Brevdo, Z. Chen, C. Citro, G. s. Corrado, A. Davis, J. Dean, M. Devin, S. Ghemawat, I. Goodfellow, A. Harp, G. Irving, M. Isard, Y. Jia, R. Jozefowicz, L. Kaiser, M. Kudlur, X. Zheng, arXiv preprint arXiv:1603.04467 2016.
[17] R. Collobert, K. Kavukcuoglu, C. Farabet, in BigLearn, Neural Information Processing Systems (NIPS) Workshop, 2011.
[18] Y. Jia, E. Shelhamer, J. Donahue, S. Karayev, J. Long, R. Girshick, S. Guadarrama, T. Darrell, Computer Science 2014.
[19] MathWorks, MATLAB Deep Learning Toolbox 2019, URL: https://www.mathworks.com/products/deep-learning.html.
[20] H. Lee, H. Kim, Magn. Reson. Med. 2019, 82, 33-48.
[21] Y. Shen, A. Bax, J. Biomol. NMR 2010, 48, 13-22.
[22] a) Y. Shen, A. J. J. o. B. N. Bax, J. Biomol. NMR 2013, 56, 227-241; b) Y. Shen, A. Bax, Methods in Molecular Biology (Clifton, N.J.) 2015, 1260, 17-32.
[23] F. H. Allen, V. J. Hoy, in International Tables for Crystallography Volume F: Crystallography ofbiological macromolecules (Eds.: M. G. Rossmann, E. Arnold), Springer, Dordrecht, 2001, pp. 663-668.
[24] S. Liu, J. Li, K. Bennett, B. Ganoe, T. Stauch, M. Head-Gordon, A. Hexemer, D. Ushizima, T. Head-Gordon, J. Phys. Chem. Lett. 2019, 10, 4558-4565.
[25] P. Klukowski, A. Gonczarek, M. J. Walczak, in 2015 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), 2015, pp. 1-8.
[26] Bruker Biospin Corporation, in European Magnetic Resonance Meeting, 2019, URL: https://www.bruker.com/fileadmin/user_upload/5-Events/2019/BBIO/EUROISMAR/Deep-Learning-Applications-in-NMR-low-res.pdf.
[27] J. C. J. Barna, E. D. Laue, M. R. Mayger, J. Skilling, S. J. P. Worrall, J. Magn. Reson. 1987, 73, 69-77.
[28] a) V. Jaravine, I. Ibraghimov, V. Yu Orekhov, Nature Methods 2006, 3, 605-607; b) X. Qu, X. Cao, D. Guo, Z. Chen, in International Society for Magnetic Resonance in Medicine 19th Scientific Meeting, 2010, pp. 3371; c) K. Kazimierczuk, J. Stanek, A. Zawadzka-Kazimierczuk, W. Koźmiński, Prog. Nucl. Mag. Res. Sp. 2010, 57, 420-434; d) K. Kazimierczuk, V. Y. Orekhov, Angew. Chem. Int. Ed. 2011, 50, 5556-5559; e) D. J. Holland, M. J. Bostock, L. F. Gladden, D. Nietlispach, Angew. Chem. Int. Ed. 2011, 50, 6548-6551; f) X. Qu, D. Guo, X. Cao, S. Cai, Z. Chen, Sensors 2011, 11, 8888-8909; g) S. G. Hyberts, A. G. Milbradt, A. B. Wagner, H. Arthanari, G. Wagner, J Biomol. NMR 2012, 52, 315-327; h) M. Mayzel, K. Kazimierczuk, V. Y. Orekhov, Chem. Commu. 2014, 50, 8947-8950; i) X. Qu, M. Mayzel, J.-F. Cai, Z. Chen, V. Orekhov, Angew. Chem. Int. Ed. 2015, 54, 852-854; j) J. Ying, F. Delaglio, D. A. Torchia, A. Bax, J. Biomol. NMR 2017, 68, 101-118.
[29] a) J. Ying, H. Lu, Q. Wei, J.-F. Cai, D. Guo, J. Wu, Z. Chen, X. Qu, IEEE Trans. Signal Proces. 2017, 65, 3702-3717; b) D. Guo, H. Lu, X. Qu, IEEE Access 2017, 5, 16033-16039; c) J. Ying, J.-F. Cai, D. Guo, G. Tang, Z. Chen, X. Qu, IEEE Trans. Signal Proces. 2018, 66, 5520-5533; d) H. Lu, X. Zhang, T. Qiu, J. Yang, J. Ying, D. Guo, Z. Chen, X. Qu, IEEE Trans. on Biomed. Eng. 2018, 65, 809-820.
[30] S. W. Provencher, Magn. Reson. Med. 1993, 30, 672-679.
[31] a) S. Williams, Prog. Nucl. Mag. Res. Sp. 1999, 35, 201; b) F. Jiru, Eur. J. Radiol. 2008, 67, 202-217.
[32] P. S. Allen, R. B. Thompson, A. H. Wilman, NMR Biomed. 1997, 10, 435-444.
[33] a) P. Luginbühl, T. Szyperski, K. Wüthrich, J. Magn. Reson. 1995, 109, 229-233; b) G. Cornilescu, F. Delaglio, A. Bax, J. Biomol. NMR 1999, 13, 289-302; c) A. Cavalli, X. Salvatella, C. M. Dobson, M. Vendruscolo, P. Natl. Acad. Sci. USA 2007, 104, 9615-9620; d) Y. Shen, O. Lange, F. Delaglio, P. Rossi, J. Aramini, G. Liu, A. Eletsky, Y. Wu, K. Singarapu, A. Lemak, A. Ignatchenko, C. Arrowsmith, T. Szyperski, G. Montelione, D. Baker, A. Bax, P. Natl. Acad. Sci. USA 2008, 105, 4685-4690; e) D. Wishart, D. Arndt, M. Berjanskii, P. Tang, J. Zhou, G. Lin, Nucleic Acids Res. 2008, 36, 496-502; f) Y. Shen, F. Delaglio, G. Cornilescu, A. Bax, J. Biomol. NMR 2009, 44, 213-223.
[34] S. Neal, A. Nip, H. Zhang, D. Wishart, J. Biomol. NMR 2003, 26, 215-240.
[35] Y. Shen, A. Bax, J. Biomol. NMR 2007, 38, 289-302.
[36] K. Kohlhoff, P. Robustelli, A. Cavalli, X. Salvatella, M. Vendruscolo, J. Am. Chem. Soc. 2009, 131, 13894-13895.
[37] J. Meiler, J. Biomol. NMR 2003, 26, 25-37.
[38] M. Williamson, C. Craven, J. Biomol. NMR 2009, 43, 131-143.
[39] G. J. Kleywegt, R. Boelens, R. Kaptein, J. Magn. Reson. 1990, 88, 601-608.
[40] B. A. Johnson, in Protein NMR Techniques (Ed.: A. K. Downing), Humana Press, NJ, 2004, pp. 313-352.
[41] C. Bartels, T.-h. Xia, M. Billeter, P. Güntert, K. Wüthrich, J. Biomol. NMR 1995, 6, 1-10.
[42] S. P. Skinner, R. H. Fogh, W. Boucher, T. J. Ragan, L. G. Mureddu, G. W. Vuister, J. Biomol. NMR 2016, 66, 111-124.
[43] Z. Liu, A. Abbas, B.-Y. Jing, X. Gao, Bioinformatics 2012, 28, 914-920.
[44] a) B. Alipanahi, X. Gao, E. Karakoc, L. Donaldson, M. Li, Bioinformatics 2009, 25, 268-275; b) S. Tikole, V. Jaravine, V. Rogov, V. Dötsch, P. Güntert, BMC Bioinformatics 2014, 15, 46.
[45] a) Y. Cheng, X. Gao, F. Liang, Genom. Proteom. Bioinf. 2014, 12, 39-47; b) J. M. Würz, P. Güntert, J. Biomol. NMR 2017, 67, 63-76.
[46] Y. Bengio, T. Deleu, N. Rahaman, R. Ke, S. Lachapelle, O. Bilaniuk, A. Goyal, C. Pal, arXiv preprint arXiv:1901.10912 2019.
[47] J. Amey, I. Kuprov, arXiv preprint arXiv:1912.01498 2019.
[48] F. Lam, Y. Li, X. Peng, IEEE Trans. Med. Imaging 2020, 39, 545-555.
[49] E. L. Ulrich, H. Akutsu, J. F. Doreleijers, Y. Harano, Y. E. Ioannidis, J. Lin, M. Livny, S. Mading, D. Maziuk, Z. Miller, E. Nakatani, C. F. Schulte, D. E. Tolmie, R. Kent Wenger, H. Yao, J. L. Markley, Nucleic Acids Res. 2007, 36, D402-D408.
[50] H. M. Berman, T. N. Bhat, P. E. Bourne, Z. Feng, G. Gilliland, H. Weissig, J. Westbrook, Nat. Struct. Mol. Biol. 2000, 7, 957-959.
[51] D. Li, R. Brüschweiler, J. Biomol. NMR 2015, 62, 403-409.
[52] M. Lundborg, G. Widmalm, Anal. Chem. 2011, 83, 1514-1517.
[53] D. Li, R. Brüschweiler, J. Biomol. NMR 2012, 54, 257-265.
[54] NMRbox, 2020, URL: https://nmrbox.org.