CloudBrain-MRS: An intelligent cloud computing platform for in vivo magnetic resonance spectroscopy
preprocessing, quantification, and analysis ( [English] )
陈晓蝶1,#, 李嘉钰1,#, 陈棣成1, 周毅荣1, 涂章仁1, 林美金2, 康泰山3, 林建忠3, 巩涛4, 朱柳红5, 周建军5, 欧阳林6, 郭杰锋7, 董继扬1, 郭迪
8, 屈小波1*
1厦门大学,电子科学系,福建等离子体与磁共振重点研究实验室,中国,厦门;
2厦门大学应用海洋物理与工程系,中国,厦门;
3厦门大学附属中山医院磁共振科,中国,厦门;.
4山东第一医科大学附属省立医院放射科,中国,山东济南;
5复旦大学附属中山医院放射科,中国,厦门;
6厦门大学医学院附属东南医院医学影像科,中国,厦门;
7厦门大学微电子与集成电路系,中国,厦门;
8厦门理工学院,计算机与信息工程学院,中国,厦门;
联系人:
quxiaobo<|at|>xmu.edu.cn
引用:
Xiaodie Chen, Jiayu Li, Dicheng Chen, Yirong Zhou, Zhangren Tu, Meijin Lin, Taishan Kang, Jianzhong Lin,
Tao Gong, Liuhong Zhu, Jianjun Zhou, Jiefeng Guo, Jiyang Dong, Di Guo, Xiaobo Qu∗, CloudBrain-MRS: An
intelligent cloud computing platform for in vivo magnetic resonance spectroscopy preprocessing, quantification,
and analysis,Journal of Magnetic Resonance, vol. 358, pp. 107601, 2023.
全文链接:https://www.sciencedirect.com/science/article/pii/S1090780723002367?via%3Dihub
摘要:
磁共振波谱(Magnetic Resonance Spectroscopy, MRS)是一种重要的临床疾病诊断方法,通过MRS可以观察代谢物的信号强度,进一步推断其浓度。虽然磁共振厂商普遍提供谱图可视化和代谢物定量等基本功能,但由于缺乏易用的处理软件或平台,MRS的临床研究推广仍受到限制。为了解决这个问题,我们开发了磁共振波谱定量分析平台CloudBrain-MRS。这是一个基于云计算的在线平台,提供强大的硬件和先进的算法。该平台只需通过浏览器即可访问,用户无需安装任何程序。CloudBrain-MRS 还集成了经典的 LCModel 模型和先进的人工智能算法,并支持对来自不同供应商的 MRS 数据进行批量预处理、量化和分析。此外,该平台还提供以下实用功能:(1)自动统计分析,寻找疾病的生物标记物;(2)经典量化算法与人工智能量化算法之间的一致性验证;(3)彩色三维可视化,方便观察单个代谢物谱。最后,健康受试者和轻度认知障碍患者的数据被用来展示该平台的功能。这是首个支持使用人工智能算法处理活体 MRS 数据的云计算平台,已共享在 MRSHub社区,提供至少两年的免费访问和服务。如果您感兴趣,请访问https://mrshub.org/software_all/#CloudBrain-MRS 或 https://csrc.xmu.edu.cn/CloudBrain.html。
关键词:
磁共振波谱、云计算、量化、数据分析、预处理
方法:
1.
背景
磁共振波谱(MRS)是一种非侵入性技术,用于量化人脑中的代谢物,以诊断各种疾病。然而,获取的 MRS
信号通常需要进行数据预处理和定量分析,以获得准确的代谢物浓度。目前,有多种开源工具可用于 MRS
信号的预处理、量化和分析。虽然这些工具提供了友好的用户界面,但仍要求用户编译源代码、下载依赖项或安装软件。此外,这些工具都不包括深度学习算法,这对当前人工智能时代的研究是一个重大限制。在过去的几十年中,已有多个 MRS
云平台用于模拟基集、从核磁共振的非采样数据中重建频谱。云平台还被应用于磁共振成像(MRI)。云计算提供了一个易于访问、灵活且可扩展的平台。用户无需担心硬件维护和管理,因此可以专注于各自专业领域的核心任务。

2.
工作流
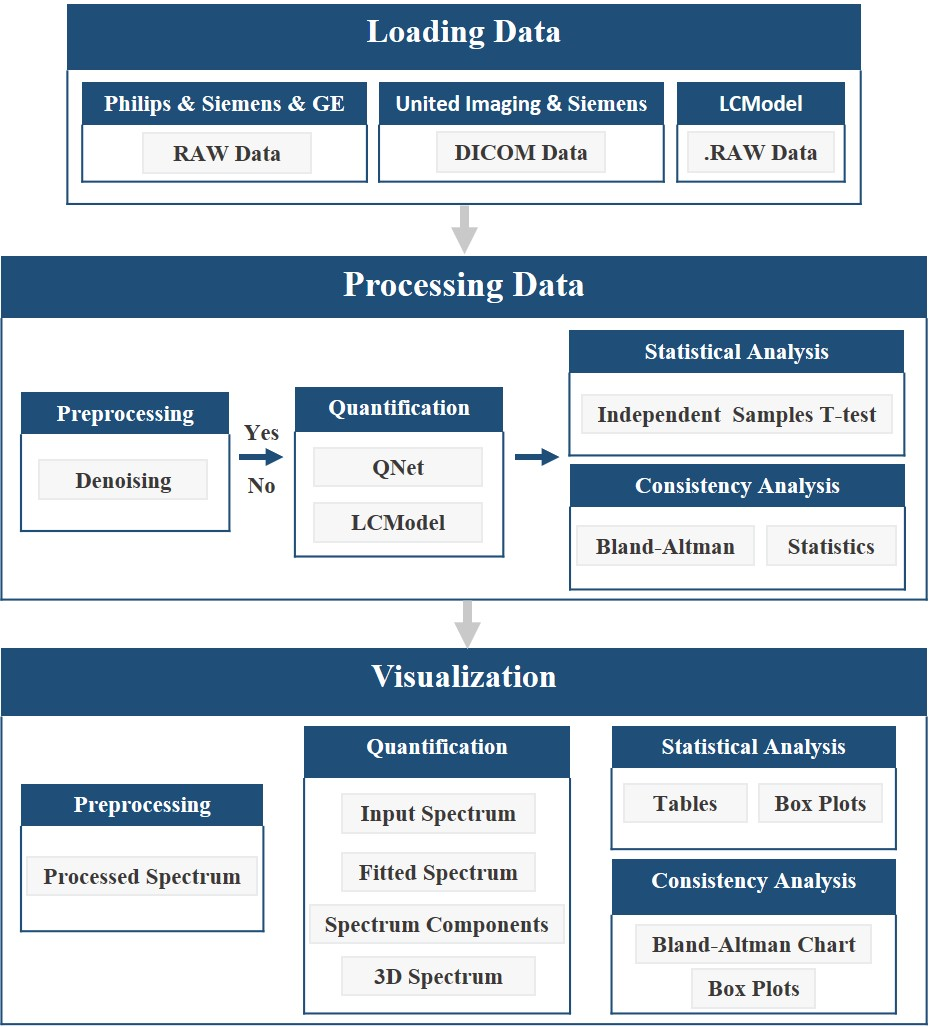
目前,该平台主要包括两个功能模块:智能量化和自动分析。用户可以注册账户或使用我们的测试账户(用户名:demo_csg,密码:csg12345678!)。主页上的使用手册也能帮助用户快速上手。CloudBrain-MRS
的工作流程如图 2 所示,详细描述如下:
(1) 加载数据和相应参数。该平台目前支持读取飞利浦、西门子和GE的 RAW 类型的数据,以及联影和西门子的 DICOM 类型的数据,还支持 LCModel 的数据格式。
(2)调用量化模型对数据进行批量或非批量量化,并保存量化结果。如果用户选择对数据进行再处理,则会在量化前进行去噪处理。
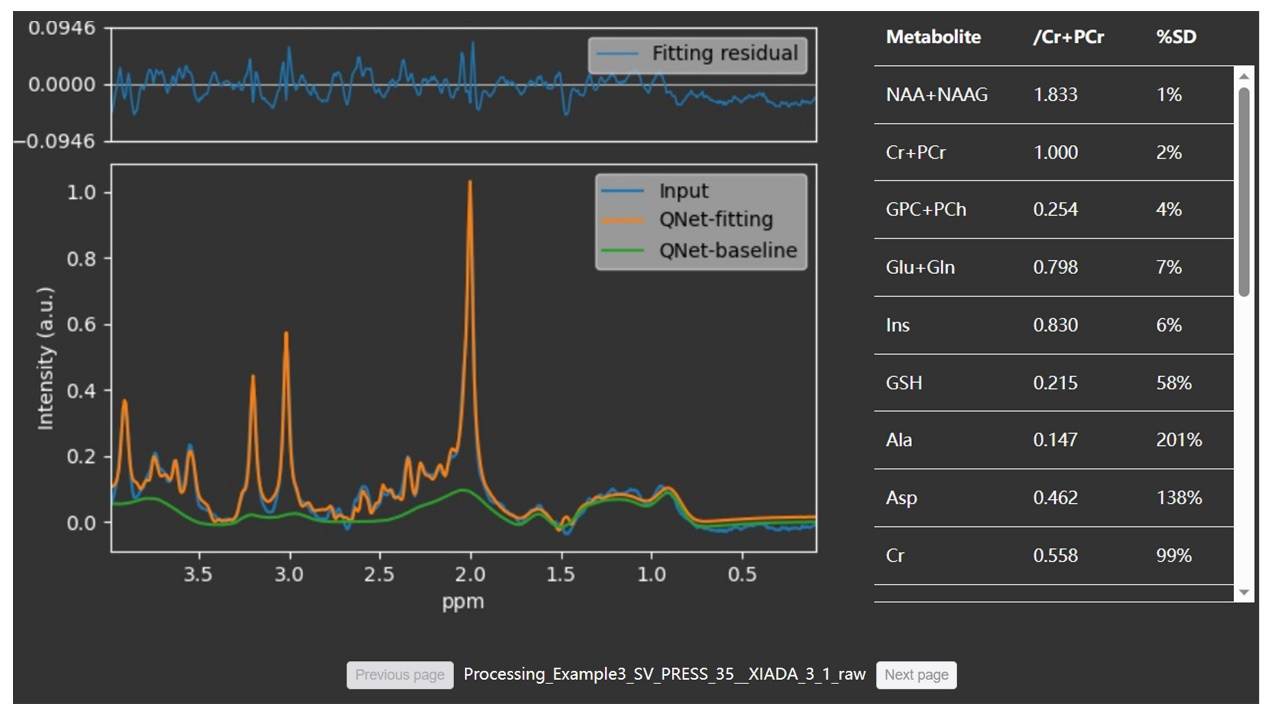
(3) 根据量化结果生成四种可视化谱图: 输入谱、整体和单个代谢物的拟合谱以及三维可视化谱。如果应用了去噪,则会显示去噪前和去噪后的谱图。
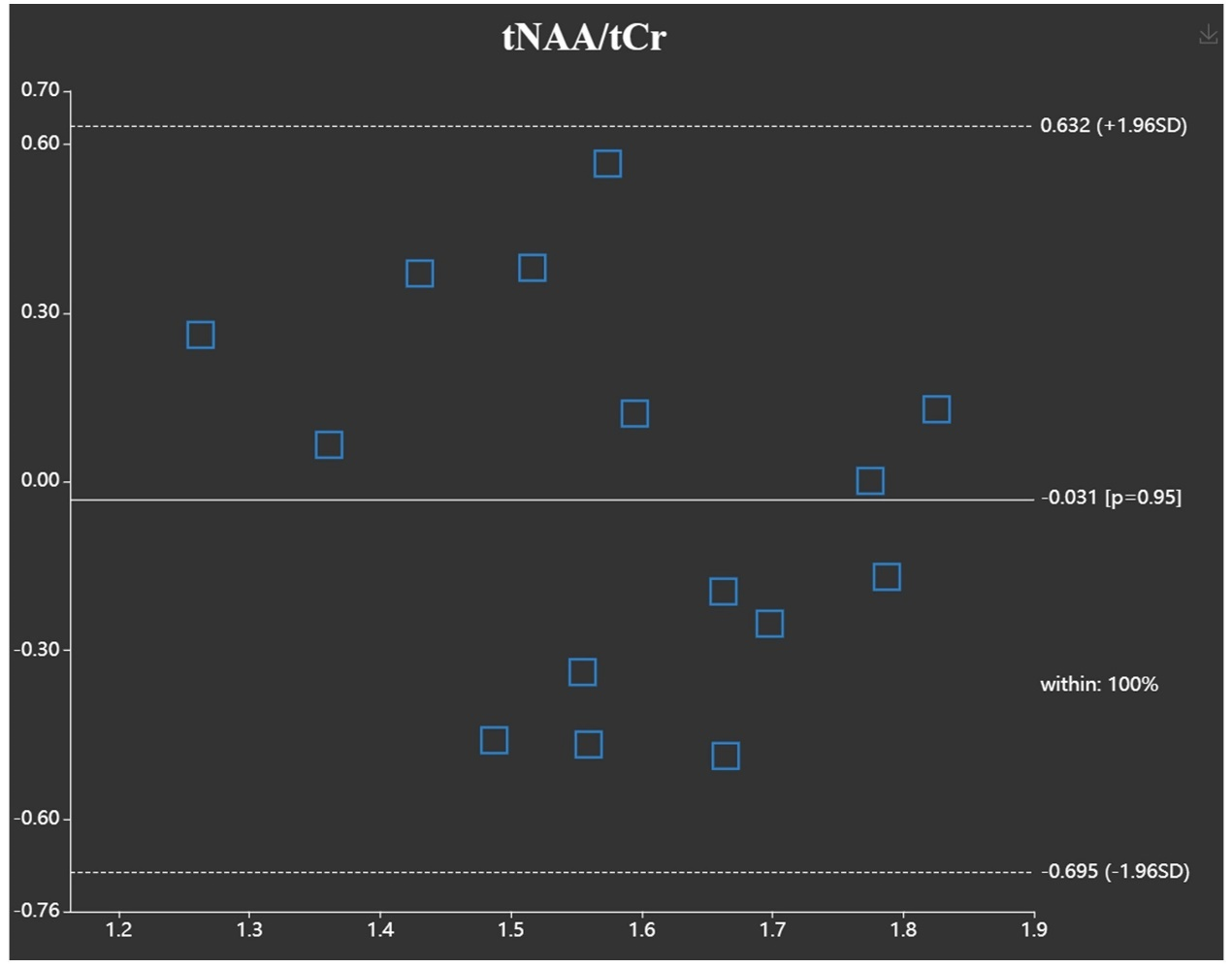
(4) 提取定量分析结果并生成相应的分析图表。统计分析将生成箱型图和三线表。一致性分析功能将生成Bland-Altman图和箱形图。
3.
系统架构
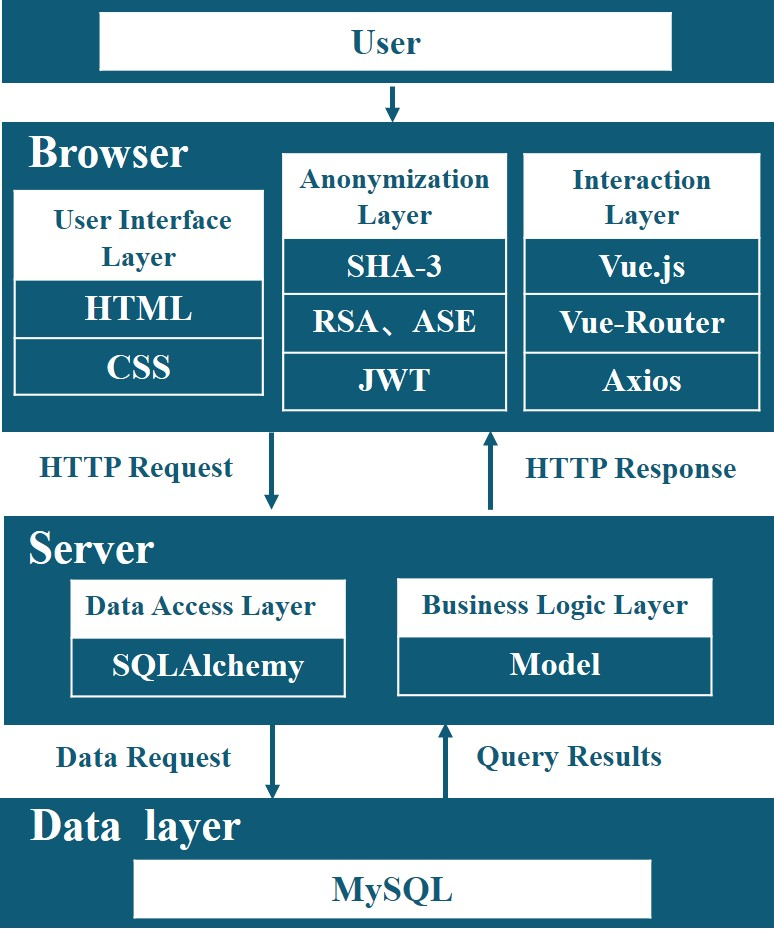
为了提高用户友好性,CloudBrain-MRS采用了浏览器/服务器(B/S)工作模式,即浏览器-请求/服务器-响应模式。系统架构可分为三个部分,即浏览器、服务器和数据库。图 3 显示了各部分之间的交互以及它们所依赖的库。
4.
系统安全和隐私
在云系统中,上传的文件会进行脱敏处理,姓名等敏感信息会被删除,但年龄和性别等数据分析所需的信息会被保留。患者隐私由浏览器端处理,不会向我们的服务器传输可识别患者身份的信息。用户有权删除数据。一旦删除,原始数据和处理过的数据都将从服务器上永久删除。
在数据安全传输方面,该平台采用加密传输和身份验证等措施,防止敏感信息被非法获取。平台采用 2048 位 Rivest-Shamir-Adleman 算法(RSA)对敏感信息进行加密后再传输。JSON
网络令牌(JWT)用于验证用户的登录状态。利用高级加密标准(ASE)算法,JWT
通过加密传输进行安全验证。在数据存储安全方面,该平台设置了允许端口白名单,对数据库实施严格的访问限制,并增加了对分布式拒绝服务(DDOS)攻击的保护。数据库和缓存中的数据使用加密存储。
5.
云平台处理结果
我们通过一些活体数据来展示该平台的实用性。


致谢:
本工作得到了国家重点研发计划(2023YFF0714200)、国家自然科学基金(62122064、61971361、62331021、62371410)、福建省自然科学基金(2023J02005、2021J011184)、厦门大学校长基金(20720220063)和厦门大学南强杰出人才计划的部分资助。感谢中国移动提供的云计算服务支持。感谢飞利浦公司的武志刚、林良杰和王家正,以及联影公司的朱家煜和张熙靖提供的技术支持。还要感谢
Stephen W. Provencher 公开了 LCModel的源码。
参考文献:
[1] G.S. Malhi, M. Valenzuela, W. Wen, P.
Sachdev, Magnetic resonance spectroscopy
and its applications in psychiatry,
Aust. N. Z. J. Psychiatry (1) (2002) 31–43.
[2] S. Behrens, H.
Laue, M. Althaus, T. Boehler, B. Kuemmerlen, H.K. Hahn, H.-O.
Peitgen, Computer assistance for MR based diagnosis of breast cancer: Present and future challenges,
Comput. Med. Imaging Graph. 31 (4–5) (2007) 236–247.
71-95, 2017.
[3] A. Gökcay, O.
Kitis, O. Ekmekci, H. Karasoy, R. Sener, Proton MR spectroscopy
in Rett syndrome,
Med. Imaging Graph. 26 (4) (2002) 271–275.
[4] R. Sener, Proton
MR spectroscopy of craniopharyngiomas, Comput. Med. Imaging
Graph.
25 (5) (2001) 417–422.
[5] J. Near, A.D.
Harris, C. Juchem, R. Kreis, M. Marjańska, G. Öz, J. Slotboom,
M. Wilson, C. Gasparovic, Preprocessing, analysis and quantification in singlevoxel magnetic resonance
spectroscopy: Experts’ consensus recommendations, NMR Biomed. 34 (5)
(2021) e4257.
[6] D.J. Drost, W.R.
Riddle, G.D. Clarke, Proton magnetic resonance spectroscopy
in the brain: Report of AAPM MR Task Group# 9,
Med. Phys. 29 (9) (2002) 2177–2197.
[7] J.-B. Poullet,
D.M. Sima, S. Van Huffel, MRS signal quantitation: A review of
time-and frequency-domain methods,
J. Magn. Reson. 195 (2) (2008) 134–144.
[8] P.K. Mandal, In
vivo proton magnetic resonance spectroscopic signal processing
for the absolute quantitation of brain metabolites,
Eur. J. Radiol. 81 (4) (2012) e653–e664.
[9] J. Near, R.
Edden, C.J. Evans, R. Paquin, A. Harris, P. Jezzard, Frequency
and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain,
Magn. Reson. Med. 73 (1) (2015) 44–50.
6, pp. 737-758, 2010.
[10] L. Vanhamme, A. van den
Boogaart, S. Van Huffel, Improved method for accurate
and efficient quantification of MRS data with use of prior knowledge, J.
Magn. Reson. 129 (1) (1997) 35–43.
[11] J.F. Jansen, W.H. Backes,
K. Nicolay, M.E. Kooi, 1H MR spectroscopy of the brain:
Absolute quantification of metabolites,
Radiology 240 (2) (2006) 318–332.
[12] J. Van der Veen, R. De
Beer, P. Luyten, D. Van Ormondt, Accurate quantification
of in vivo 31P NMR signals using the variable projection method and prior knowledge, Magn. Reson. Med. 6 (1) (1988) 92–98.
[13] S.W. Provencher, Estimation
of metabolite concentrations from localized in vivo
proton NMR spectra,
Magn. Reson. Med. 30 (6) (1993) 672–679.
[14] H. Ratiney, M. Sdika, Y.
Coenradie, S. Cavassila, D.V. Ormondt, D. GraveronDemilly, Time-domain semi-parametric estimation based on a
metabolite basis set,
NMR Biomed. 18 (1) (2005) 1–13.
[15] J.-B. Poullet, D.M. Sima,
A.W. Simonetti, B. De Neuter, L. Vanhamme, P.
Lemmerling, S. Van Huffel, An automated quantitation of short echo time MRS spectra in an open source software
environment: AQSES,
NMR Biomed. 20 (5) (2007) 493–504.
[16] S. Tapper, M. Mikkelsen,
B.E. Dewey, H.J. Zöllner, S.C. Hui, G. Oeltzschner, R.A.
Edden, Frequency and phase correction of J-difference edited MR spectra using deep learning, Magn. Reson. Med. 85 (4) (2021) 1755–1765.
[17] D.J. Ma, H.A.-M. Le, Y. Ye,
A.F. Laine, J.A. Lieberman, D.L. Rothman, S.A. Small,
J. Guo, MR spectroscopy frequency and phase correction using convolutional neural networks,
Magn. Reson. Med. 87 (4) (2022) 1700–1710.
[18] D. Chen, W. Hu, H. Liu, Y.
Zhou, T. Qiu, Y. Huang, Z. Wang, M. Lin, L. Lin,
Z. Wu, J. Wang, H. Chen, X. Chen, G. Yan, D. Guo, J. Lin, X. Qu, Magnetic resonance spectroscopy deep learning
denoising using few in vivo data, IEEE Trans. Comput. Imaging 9 (2023)
448–458.
[19] J. Jang, H.H. Lee, J.-A.
Park, H. Kim, Unsupervised anomaly detection using
generative adversarial networks in 1H-MRS of the brain,
J. Magn. Reson. 325 (2021) 106936.
[20] N. Hatami, M. Sdika, H.
Ratiney, Magnetic resonance spectroscopy quantification
using deep learning, in: International Conference on Medical Image
Computing and Computer Assisted Intervention, 2018, pp. 467–475.
[21] M. Chandler, C. Jenkins, S.
Shermer, F. Langbein, MRSNet: metabolite quantification from edited magnetic resonance spectra with
convolutional neural networks, 2019, arXiv preprint arXiv:1909.03836.
[22] S.S. Gurbani, S. Sheriff,
A.A. Maudsley, H. Shim, L.A. Cooper, Incorporation of a
spectral model in a convolutional neural network for accelerated spectral fitting, Magn. Reson. Med. 81 (5) (2019) 3346–3357.
[23] H.H. Lee, H. Kim, Intact
metabolite spectrum mining by deep learning in proton
magnetic resonance spectroscopy of the brain,
Magn. Reson. Med. 82 (1) (2019) 33–48.
[24] D. Chen, M. Lin, H. Liu, J.
Li, Y. Zhou, T. Kang, L. Lin, Z. Wu, J. Wang, J. Li,
J. Lin, X. Chen, D. Guo, X. Qu, Magnetic resonance spectroscopy quantification
aided by deep estimations of imperfection factors and overall macromolecular
signal, 2023, arXiv preprint arXiv:2306.09681.
[25] A. Naressi, C. Couturier,
J. Devos, M. Janssen, C. Mangeat, R.d. Beer, D.
Graveron-Demilly, Java-based graphical user interface for the MRUI quantitation package, Magn. Reson. Mater. Phys. Biol. Med. 12 (2001) 141–152.
[26] D. Stefan, F. Di Cesare, A.
Andrasescu, E. Popa, A. Lazariev, E. Vescovo, O.
Strbak, S. Williams, Z. Starcuk, M. Cabanas, et al., Quantitation of magnetic resonance spectroscopy signals:
The jMRUI software package, Meas. Sci. Technol. 20 (10) (2009) 104035.
[27] M. Wilson, G. Reynolds,
R.A. Kauppinen, T.N. Arvanitis, A.C. Peet, A constrained
least-squares approach to the automated quantitation of in vivo 1H magnetic resonance spectroscopy data,
Magn. Reson. Med. 65 (1) (2011) 1–12.
[28] W.T. Clarke, C.J. Stagg, S.
Jbabdi, FSL-MRS: An end-to-end spectroscopy analysis
package,
Magn. Reson. Med. 85 (6) (2021) 2950–2964.
[29] G. Oeltzschner, H.J.
Zöllner, S.C. Hui, M. Mikkelsen, M.G. Saleh, S. Tapper, R.A.
Edden, Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data,
J. Neurosci. Methods
343 (2020) 108827.
[30] R.A. Edden, N.A. Puts, A.D.
Harris, P.B. Barker, C.J. Evans, Gannet: A batchprocessing tool for the quantitative analysis of
gamma-aminobutyric acid–edited MR spectroscopy spectra, J. Magn. Reson.
Imaging 40 (6) (2014) 1445–1452.
1413-1457, 2004.
[31] R. Simpson, G.A. Devenyi,
P. Jezzard, T.J. Hennessy, J. Near, Advanced processing and simulation of MRS data using the FID appliance
(FID-A)—an open source, MATLAB-based toolkit, Magn. Reson. Med. 77 (1)
(2017) 23–33.
[32] S.C. Hui, M.G. Saleh, H.J.
Zöllner, G. Oeltzschner, H. Fan, Y. Li, Y. Song, H.
Jiang, J. Near, H. Lu, et al., MRSCloud: A cloud-based MRS tool for basis set simulation, Magn. Reson. Med. 88 (5) (2022) 1994–2004.
[33] Z. Wang, D. Guo, Z. Tu, Y.
Huang, Y. Zhou, J. Wang, L. Feng, D. Lin, Y.
You, T. Agback, et al., A sparse model-inspired deep thresholding network for exponential signal
reconstruction-application in fast biological spectroscopy,
IEEE Trans. Neural Netw. Learn. Syst. (2022).
[34] S. Gurbani, B. Weinberg, L.
Cooper, E. Mellon, E. Schreibmann, S. Sheriff, A.
Maudsley, M. Goryawala, H.-K. Shu, H. Shim, The brain imaging collaboration suite (BrICS): A cloud platform for
integrating whole-brain spectroscopic MRI into the radiation therapy planning workflow,
Tomogr.
5 (1) (2019) 184–191.
[35] Q. Yang, Z. Wang, K. Guo,
C. Cai, X. Qu, Physics-driven synthetic data learning
for biomedical magnetic resonance: The imaging physics-based data synthesis paradigm for artificial
intelligence,
IEEE Signal Process. Mag. 40 (2) (2023) 129–140.
[36] Y. Zhou, C. Qian, J. Li, Z.
Wang, Y. Hu, B. Qu, L. Zhu, J. Zhou, T. Kang, J.
Lin, Q. Hong, J. Dong, D. Guo, X. Qu, CloudBrain-ReconAI: An online platform for MRI reconstruction and image
quality evaluation, 2022, arXiv preprint arXiv: 2212.01878.
[37] H. Xue, S. Inati, T.S.
Sørensen, P. Kellman, M.S. Hansen, Distributed MRI
reconstruction using gadgetron-based cloud computing,
Magn. Reson. Med. 73 (3) (2015) 1015–1025.
[38] S. Mori, D. Wu, C.
Ceritoglu, Y. Li, A. Kolasny, M.A. Vaillant, A.V. Faria, K.
Oishi, M.I. Miller, MRICloud: delivering high-throughput MRI neuroinformatics as cloud-based software as a
service,
Comput. Sci. Eng. 18 (5) (2016) 21–35.
[39] F. Milletari, J. Frei, M.
Aboulatta, G. Vivar, S.-A. Ahmadi, Cloud deployment of
high-resolution medical image analysis with TOMAAT, IEEE J. Biomed. Health
Inform. 23 (3) (2018) 969–977.
[40] C.G. Xanthis, A.H. Aletras,
CoreMRI: A high-performance, publicly available MR
simulation platform on the cloud,
PLoS One 14 (5) (2019) e0216594.
[41] H. Wang, L. Tan, H.-F.
Wang, Y. Liu, R.-H. Yin, W.-Y. Wang, X.-L. Chang,
T. Jiang, J.-T. Yu, Magnetic resonance spectroscopy in Alzheimer’s disease: Systematic review and meta-analysis,
J. Alzheimers Dis. 46 (4) (2015) 1049–1070.
[42] A. Kherchouche, O.
Ben-Ahmed, C. Guillevin, B. Tremblais, A. Julian, C.
Fernandez-Maloigne, R. Guillevin, Attention-guided neural network for early dementia detection using MRS data,
Comput. Med. Imaging Graph. (2022) 102074.
[43] P. Lazen, P.L. Cardoso, S.
Sharma, C. Cadrien, T. Roetzer-Pejrimovsky, J. Furtner,
B. Strasser, L. Hingerl, A. Lipka, M. Preusser, W. Marik, W. Bogner, G. Widhalm,
K. Rössler, S. Trattnig, G. Hangel, A comparison of 7 Tesla MR spectroscopic
imaging and 3 Tesla MR fingerprinting for tumor localization in glioma patients,
2023, arXiv preprint arXiv:2304.05254.
[44] S. Nakae, K. Murayama, H.
Sasaki, M. Kumon, Y. Nishiyama, S. Ohba, K.
Adachi, S. Nagahisa, T. Hayashi, J. Inamasu, et al., Prediction of genetic subgroups in adult supra tentorial
gliomas by pre-and intraoperative parameters,
J. Neurooncol. 131 (2017) 403–412.
[45] T. Kazda, M. Bulik, P.
Pospisil, R. Lakomy, M. Smrcka, P. Slampa, R. Jancalek,
Advanced MRI increases the diagnostic accuracy of recurrent glioblastoma: Single institution thresholds and
validation of MR spectroscopy and diffusion weighted MR imaging,
NeuroImage Clin. 11 (2016) 316–321.
[46] L. Mazuel, C. Chassain, B.
Jean, B. Pereira, A. Cladière, C. Speziale, F. Durif,
Proton MR spectroscopy for diagnosis and evaluation of treatment efficacy in Parkinson disease,
Radiology 278 (2) (2016) 505–513.
[47] A. Flamez, W. Wiels, P. Van
Schuerbeek, J. De Mey, J. De Keyser, C. Baeken,
The influence of one session of low frequency rTMS on pre-supplementary motor area metabolites in late stage
Parkinson’s disease,
Clin. Neurophysiol. 130 (8) (2019) 1292–1298.
[48] H.-Y. Kim, Statistical
notes for clinical researchers: the independent samples
t-test,
Restor. Dent. Endod. 44 (3) (2019).
[49] M. Terpstra, I. Cheong, T.
Lyu, D.K. Deelchand, U.E. Emir, P. Bednařík, L.E.
Eberly, G. Öz, Test-retest reproducibility of neurochemical profiles with shortecho, single-voxel MR
spectroscopy at 3T and 7T,
Magn. Reson. Med. 76 (4) (2016) 1083–1091.
[50] I. Tkáč, G. Öz, G. Adriany,
K. Uğurbil, R. Gruetter, In vivo 1H NMR spectroscopy
of the human brain at high magnetic fields: Metabolite quantification at 4T vs. 7T,
Magn. Reson. Med. 62 (4) (2009) 868–879.
[51] V. Govindaraju, K. Young,
A.A. Maudsley, Proton NMR chemical shifts and
coupling constants for brain metabolites,
NMR Biomed. 13 (3) (2000) 129–153.
[52] R. Kreis, The trouble with
quality filtering based on relative C ramér-R ao lower
bounds,
Magn. Reson. Med. 75 (1) (2016) 15–18.
[53] A. Metastasio, P. Rinaldi,
R. Tarducci, E. Mariani, F.T. Feliziani, A. Cherubini,
G.P. Pelliccioli, G. Gobbi, U. Senin, P. Mecocci, Conversion of MCI to dementia: Role of proton magnetic
resonance spectroscopy,
Neurobiol. Aging 27 (7) (2006) 926–932.
[54] K. Kantarci, Proton MRS in
mild cognitive impairment,
J. Magn. Reson. Imaging 37 (4) (2013) 770–777.
[55] S. Tumati, S. Martens, A.
Aleman, Magnetic resonance spectroscopy in mild
cognitive impairment: Systematic review and meta-analysis,
Neurosci. Biobehav. Rev. 37 (10) (2013) 2571–2586.