CloudBrain-MRS: An intelligent cloud computing platform for in vivo magnetic resonance spectroscopy
preprocessing, quantification, and analysis ( [Chinese] )
Xiaodie Chena,1, Jiayu Lia,1, Dicheng Chena, Yirong Zhoua, Zhangren Tua, Meijin Linb, Taishan Kangc, Jianzhong Linc, Tao Gongd, Liuhong Zhue, Jianjun Zhoue, Lin Ou-yangf, Jiefeng Guog, Jiyang Donga, Di Guo
h, Xiaobo Qua,*
aDepartment of Electronic Science, Fujian Provincial
Key Laboratory of Plasma and Magnetic Resonance, Xiamen University, Xiamen, China
bDepartment of Applied Marine Physics & Engineering,
Xiamen University, Xiamen, China
cDepartment of Radiology, Zhongshan Hospital
Affiliated to Xiamen University, Xiamen, China
dDepartments of Radiology, Shandong Provincial
Hospital Affiliated to Shandong First Medical University, Jinan, Shandong, China
eDepartment of Radiology, Zhongshan Hospital
(Xiamen), Fudan University, Xiamen, China
fDepartment of Medical Imaging of Southeast Hospital,
Medical College of Xiamen University, Xiamen, China
gDepartment of Microelectronics and Integrated
Circuit, Xiamen University, Xiamen, China
hSchool of Computer and Information Engineering,
Xiamen University of Technology, Xiamen, China
Concat:
quxiaobo<|at|>xmu.edu.cn
Citation:
Xiaodie Chen, Jiayu Li, Dicheng Chen, Yirong Zhou, Zhangren Tu, Meijin Lin, Taishan Kang, Jianzhong Lin,
Tao Gong, Liuhong Zhu, Jianjun Zhou, Jiefeng Guo, Jiyang Dong, Di Guo, Xiaobo Qu∗, CloudBrain-MRS: An
intelligent cloud computing platform for in vivo magnetic resonance spectroscopy preprocessing, quantification,
and analysis, Journal of Magnetic Resonance, vol. 358, pp. 107601, 2023.
Access to full text:https://www.sciencedirect.com/science/article/pii/S1090780723002367?via%3Dihub
Abstract:
Magnetic resonance spectroscopy (MRS) is an important clinical imaging method for diagnosis of diseases. MRS spectrum is used to observe the signal intensity of metabolites or further infer their concentrations. Although the magnetic resonance vendors commonly provide basic functions of spectrum plots and metabolite quantification, the spread of clinical research of MRS is still limited due to the lack of easy-to-use processing software or platform. To address this issue, we have developed CloudBrain-MRS, a cloud-based online platform that provides powerful hardware and advanced algorithms. The platform can be accessed simply through a web browser, without the need of any program installation on the user side. CloudBrain-MRS also integrates the classic LCModel and advanced artificial intelligence algorithms and supports batch preprocessing, quantification, and analysis of MRS data from different vendors. Additionally, the platform offers useful functions: (1) Automatically statistical analysis to find biomarkers for diseases; (2) Consistency verification between the classic and artificial intelligence quantification algorithms; (3) Colorful three-dimensional visualization for easy observation of individual metabolite spectrum. Last, data of both healthy subjects and patients with mild cognitive impairment are used to demonstrate the functions of the platform. To the best of our knowledge, this is the first cloud computing platform for in vivo MRS with artificial intelligence processing. We have shared our cloud platform at MRSHub, providing at least two years of free access and service. If you are interested, please visit https://mrshub.org/software_all/#CloudBrain-MRS or https://csrc.xmu.edu.cn/CloudBrain.html.
KEYWORDS:
Magnetic resonance spectroscopy, Cloud computing, Quantification, Data
analysis, Preprocessing
Methods:
1.
Background
Magnetic resonance spectroscopy (MRS) is a non-invasive technique used to
quantify metabolites in the human brain to diagnose various diseases. However, the acquired MRS signals
typically require data preprocessing and quantitative analysis to obtain accurate metabolite concentrations.
Currently, there are various open-source tools available for preprocessing, quantification, and analysis of MRS
signals. While these tools provide a user-friendly interface, they still require users to compile source code,
download dependencies, or install the software. Furthermore, none of these tools include deep learning
algorithms, which is a significant limitation for the current research in the era of artificial intelligence.
There is a strong need for a userfriendly system that can enable biomedical researchers and clinical
radiologists to apply these advanced algorithms effectively to clinical research. In the past few decades, there
have been several MRS cloud platforms. Cloud platforms also have been applied in magnetic resonance imaging
(MRI). Cloud computing provides an easily accessible, flexible, and scalable platform. Users need not worry
about hardware maintenance and management, and thus, they can focus on the core tasks of their field of
expertise. In this paper, we present our cloud computing platform for MRS with the entire processing and
postprocessing procedure.

2.
Workflow summary
Currently, the
platform mainly
contains two functional modules: Intelligent quantification and automatic analysis. Users can register an
account or use our demo account (username: demo_csg, password: csg12345678!). The manual on the homepage also
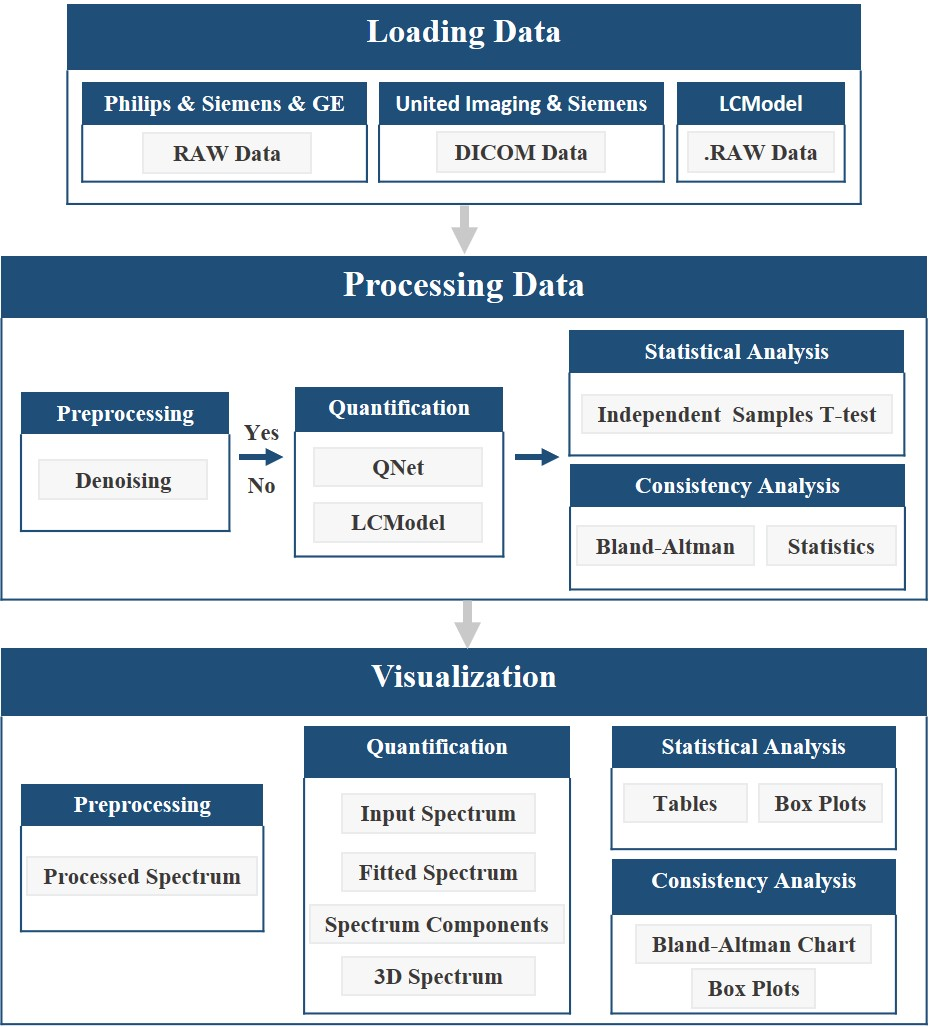
can help users to get started quickly. The workflow of CloudBrain-MRS is illustrated in Fig. 2 and can be
described in detail as follows:
(1) Load the data and the corresponding parameters. The platform currently supports reading RAW data from
Philips, Siemens, and GE, DICOM data from United Imaging and Siemens, and also supports LCModel’s data
format.
(2) Invoke the quantification model to quantify the data either in batch or not, and save the quantification
results. If a user chooses to preprocess the data, denoising will be performed before quantification.
(3) Generate four types of visual spectra based on the quantification results: Inputted spectrum, fitted spectra
of overall and individual metabolites, and 3D visualized spectrum. If denoising is applied, both the spectra
before and after denoising will be shown.
(4) Extract the quantitative results for analysis and generate the corresponding analysis charts. Statistical
analysis will generate box plots and trilinear tables. The function of ‘‘Consistency Analysis’’ will generate
Bland-Altman charts and box plots.
3.
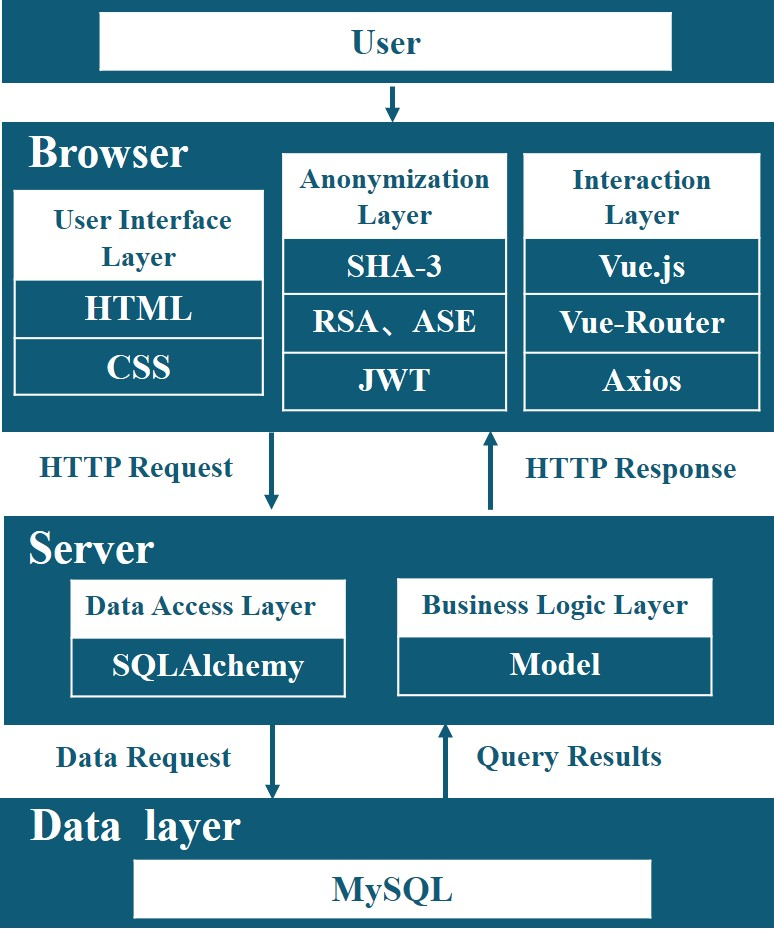
Architecture of the system
To
enhance user-friendliness, CloudBrain-MRS adopts the browser/ service (B/S) working model, which stands for a
browser-request/ server-response model. The system architecture can be divided into three parts, namely browser,
server, and database. Fig. 3 displays the interactions between each part and the libraries they depend on.
4.
Security and privacy in the system
In the cloud system, uploaded files are desensitized,
and sensitive information such as names are deleted, but the information needed for data analysis such as age
and gender is retained. Patient privacy is handled at the browser and no patient-identifiable information is
transmitted to our server. Users have the right to delete data. Once deleted, both the original and processed
data are permanently deleted from the server.
For data secure transmission, CloudBrain-MRS adopts measures such as encrypted transmission
and identity
authentication to prevent sensitive information from being illegally obtained. The platform uses the 2048-bit
Rivest–Shamir–Adleman (RSA) algorithm to encrypt sensitive information before transmission. JSON Web Token (JWT)
is utilized for validating the user’s login status. With the Advanced Encryption Standard (ASE) algorithm, JWT
is transmitted with encryption for secure authentication.
For data storage security, the platform sets up a white list of allowed ports to impose
strict access
restrictions on the database and adds protection against distributed denial of service (DDOS) attacks. Data in
the database and cache are stored using encrypted storage.
5.
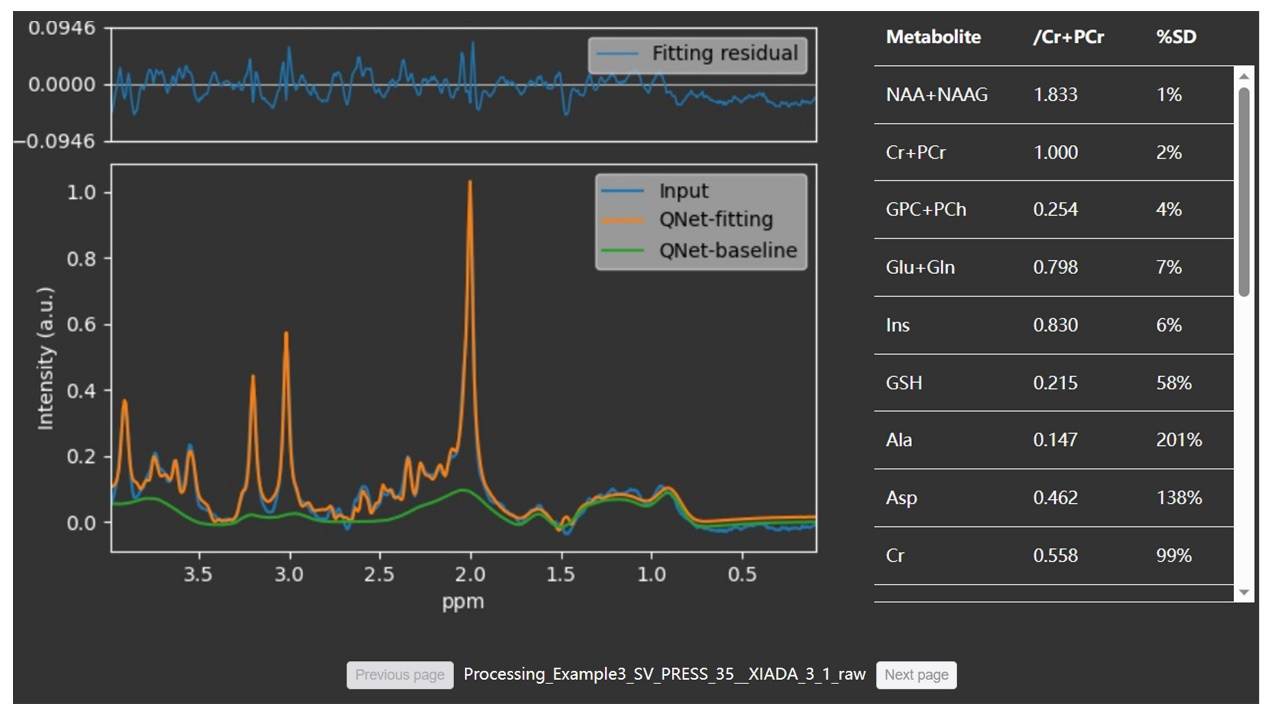
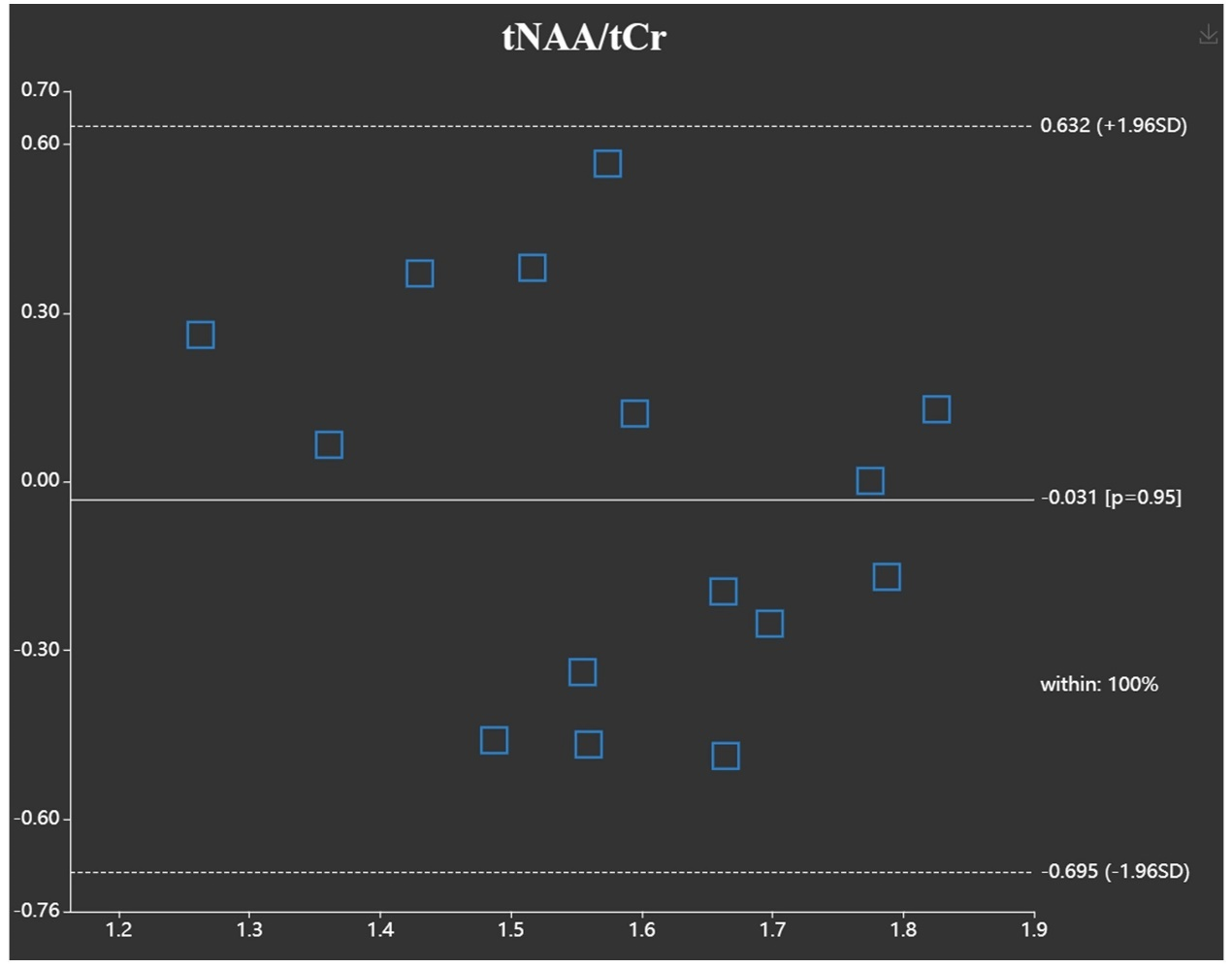
Resluts
We
demonstrate the usefulness of the platform with some in vivo data.


Acknowledgments:
This work was partially supported by the National Key Research and
Development Program,
China (2023YFF0714200), the National Natural Science Foundation of China (62122064, 61971361, 62331021,
62371410), the Natural Science Foundation of Fujian Province of China (2023J02005, 2021J011184), the President
Fund of Xiamen University, China (20720220063), and Nanqiang Outstanding Talent Program of Xiamen University,
China. The authors thank China Mobile for providing cloud computing services support. The authors thank Zhigang
Wu, Liangjie Lin, and Jiazheng Wang from Philips and Jiayu Zhu and Xijing Zhang from United Imaging for
technical support. The authors also thank Stephen W. Provencher for making LCModel public.
Reference:
[1] G.S. Malhi, M. Valenzuela, W. Wen, P.
Sachdev, Magnetic resonance spectroscopy
and its applications in psychiatry,
Aust. N. Z. J. Psychiatry (1) (2002) 31–43.
[2] S. Behrens, H.
Laue, M. Althaus, T. Boehler, B. Kuemmerlen, H.K. Hahn, H.-O.
Peitgen, Computer assistance for MR based diagnosis of breast cancer: Present and future challenges,
Comput. Med. Imaging Graph. 31 (4–5) (2007) 236–247.
71-95, 2017.
[3] A. Gökcay, O.
Kitis, O. Ekmekci, H. Karasoy, R. Sener, Proton MR spectroscopy
in Rett syndrome,
Med. Imaging Graph. 26 (4) (2002) 271–275.
[4] R. Sener, Proton
MR spectroscopy of craniopharyngiomas, Comput. Med. Imaging
Graph.
25 (5) (2001) 417–422.
[5] J. Near, A.D.
Harris, C. Juchem, R. Kreis, M. Marjańska, G. Öz, J. Slotboom,
M. Wilson, C. Gasparovic, Preprocessing, analysis and quantification in singlevoxel magnetic resonance
spectroscopy: Experts’ consensus recommendations, NMR Biomed. 34 (5)
(2021) e4257.
[6] D.J. Drost, W.R.
Riddle, G.D. Clarke, Proton magnetic resonance spectroscopy
in the brain: Report of AAPM MR Task Group# 9,
Med. Phys. 29 (9) (2002) 2177–2197.
[7] J.-B. Poullet,
D.M. Sima, S. Van Huffel, MRS signal quantitation: A review of
time-and frequency-domain methods,
J. Magn. Reson. 195 (2) (2008) 134–144.
[8] P.K. Mandal, In
vivo proton magnetic resonance spectroscopic signal processing
for the absolute quantitation of brain metabolites,
Eur. J. Radiol. 81 (4) (2012) e653–e664.
[9] J. Near, R.
Edden, C.J. Evans, R. Paquin, A. Harris, P. Jezzard, Frequency
and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain,
Magn. Reson. Med. 73 (1) (2015) 44–50.
6, pp. 737-758, 2010.
[10] L. Vanhamme, A. van den
Boogaart, S. Van Huffel, Improved method for accurate
and efficient quantification of MRS data with use of prior knowledge, J.
Magn. Reson. 129 (1) (1997) 35–43.
[11] J.F. Jansen, W.H. Backes,
K. Nicolay, M.E. Kooi, 1H MR spectroscopy of the brain:
Absolute quantification of metabolites,
Radiology 240 (2) (2006) 318–332.
[12] J. Van der Veen, R. De
Beer, P. Luyten, D. Van Ormondt, Accurate quantification
of in vivo 31P NMR signals using the variable projection method and prior knowledge, Magn. Reson. Med. 6 (1) (1988) 92–98.
[13] S.W. Provencher, Estimation
of metabolite concentrations from localized in vivo
proton NMR spectra,
Magn. Reson. Med. 30 (6) (1993) 672–679.
[14] H. Ratiney, M. Sdika, Y.
Coenradie, S. Cavassila, D.V. Ormondt, D. GraveronDemilly, Time-domain semi-parametric estimation based on a
metabolite basis set,
NMR Biomed. 18 (1) (2005) 1–13.
[15] J.-B. Poullet, D.M. Sima,
A.W. Simonetti, B. De Neuter, L. Vanhamme, P.
Lemmerling, S. Van Huffel, An automated quantitation of short echo time MRS spectra in an open source software
environment: AQSES,
NMR Biomed. 20 (5) (2007) 493–504.
[16] S. Tapper, M. Mikkelsen,
B.E. Dewey, H.J. Zöllner, S.C. Hui, G. Oeltzschner, R.A.
Edden, Frequency and phase correction of J-difference edited MR spectra using deep learning, Magn. Reson. Med. 85 (4) (2021) 1755–1765.
[17] D.J. Ma, H.A.-M. Le, Y. Ye,
A.F. Laine, J.A. Lieberman, D.L. Rothman, S.A. Small,
J. Guo, MR spectroscopy frequency and phase correction using convolutional neural networks,
Magn. Reson. Med. 87 (4) (2022) 1700–1710.
[18] D. Chen, W. Hu, H. Liu, Y.
Zhou, T. Qiu, Y. Huang, Z. Wang, M. Lin, L. Lin,
Z. Wu, J. Wang, H. Chen, X. Chen, G. Yan, D. Guo, J. Lin, X. Qu, Magnetic resonance spectroscopy deep learning
denoising using few in vivo data, IEEE Trans. Comput. Imaging 9 (2023)
448–458.
[19] J. Jang, H.H. Lee, J.-A.
Park, H. Kim, Unsupervised anomaly detection using
generative adversarial networks in 1H-MRS of the brain,
J. Magn. Reson. 325 (2021) 106936.
[20] N. Hatami, M. Sdika, H.
Ratiney, Magnetic resonance spectroscopy quantification
using deep learning, in: International Conference on Medical Image
Computing and Computer Assisted Intervention, 2018, pp. 467–475.
[21] M. Chandler, C. Jenkins, S.
Shermer, F. Langbein, MRSNet: metabolite quantification from edited magnetic resonance spectra with
convolutional neural networks, 2019, arXiv preprint arXiv:1909.03836.
[22] S.S. Gurbani, S. Sheriff,
A.A. Maudsley, H. Shim, L.A. Cooper, Incorporation of a
spectral model in a convolutional neural network for accelerated spectral fitting, Magn. Reson. Med. 81 (5) (2019) 3346–3357.
[23] H.H. Lee, H. Kim, Intact
metabolite spectrum mining by deep learning in proton
magnetic resonance spectroscopy of the brain,
Magn. Reson. Med. 82 (1) (2019) 33–48.
[24] D. Chen, M. Lin, H. Liu, J.
Li, Y. Zhou, T. Kang, L. Lin, Z. Wu, J. Wang, J. Li,
J. Lin, X. Chen, D. Guo, X. Qu, Magnetic resonance spectroscopy quantification
aided by deep estimations of imperfection factors and overall macromolecular
signal, 2023, arXiv preprint arXiv:2306.09681.
[25] A. Naressi, C. Couturier,
J. Devos, M. Janssen, C. Mangeat, R.d. Beer, D.
Graveron-Demilly, Java-based graphical user interface for the MRUI quantitation package, Magn. Reson. Mater. Phys. Biol. Med. 12 (2001) 141–152.
[26] D. Stefan, F. Di Cesare, A.
Andrasescu, E. Popa, A. Lazariev, E. Vescovo, O.
Strbak, S. Williams, Z. Starcuk, M. Cabanas, et al., Quantitation of magnetic resonance spectroscopy signals:
The jMRUI software package, Meas. Sci. Technol. 20 (10) (2009) 104035.
[27] M. Wilson, G. Reynolds,
R.A. Kauppinen, T.N. Arvanitis, A.C. Peet, A constrained
least-squares approach to the automated quantitation of in vivo 1H magnetic resonance spectroscopy data,
Magn. Reson. Med. 65 (1) (2011) 1–12.
[28] W.T. Clarke, C.J. Stagg, S.
Jbabdi, FSL-MRS: An end-to-end spectroscopy analysis
package,
Magn. Reson. Med. 85 (6) (2021) 2950–2964.
[29] G. Oeltzschner, H.J.
Zöllner, S.C. Hui, M. Mikkelsen, M.G. Saleh, S. Tapper, R.A.
Edden, Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data,
J. Neurosci. Methods
343 (2020) 108827.
[30] R.A. Edden, N.A. Puts, A.D.
Harris, P.B. Barker, C.J. Evans, Gannet: A batchprocessing tool for the quantitative analysis of
gamma-aminobutyric acid–edited MR spectroscopy spectra, J. Magn. Reson.
Imaging 40 (6) (2014) 1445–1452.
1413-1457, 2004.
[31] R. Simpson, G.A. Devenyi,
P. Jezzard, T.J. Hennessy, J. Near, Advanced processing and simulation of MRS data using the FID appliance
(FID-A)—an open source, MATLAB-based toolkit, Magn. Reson. Med. 77 (1)
(2017) 23–33.
[32] S.C. Hui, M.G. Saleh, H.J.
Zöllner, G. Oeltzschner, H. Fan, Y. Li, Y. Song, H.
Jiang, J. Near, H. Lu, et al., MRSCloud: A cloud-based MRS tool for basis set simulation, Magn. Reson. Med. 88 (5) (2022) 1994–2004.
[33] Z. Wang, D. Guo, Z. Tu, Y.
Huang, Y. Zhou, J. Wang, L. Feng, D. Lin, Y.
You, T. Agback, et al., A sparse model-inspired deep thresholding network for exponential signal
reconstruction-application in fast biological spectroscopy,
IEEE Trans. Neural Netw. Learn. Syst. (2022).
[34] S. Gurbani, B. Weinberg, L.
Cooper, E. Mellon, E. Schreibmann, S. Sheriff, A.
Maudsley, M. Goryawala, H.-K. Shu, H. Shim, The brain imaging collaboration suite (BrICS): A cloud platform for
integrating whole-brain spectroscopic MRI into the radiation therapy planning workflow,
Tomogr.
5 (1) (2019) 184–191.
[35] Q. Yang, Z. Wang, K. Guo,
C. Cai, X. Qu, Physics-driven synthetic data learning
for biomedical magnetic resonance: The imaging physics-based data synthesis paradigm for artificial
intelligence,
IEEE Signal Process. Mag. 40 (2) (2023) 129–140.
[36] Y. Zhou, C. Qian, J. Li, Z.
Wang, Y. Hu, B. Qu, L. Zhu, J. Zhou, T. Kang, J.
Lin, Q. Hong, J. Dong, D. Guo, X. Qu, CloudBrain-ReconAI: An online platform for MRI reconstruction and image
quality evaluation, 2022, arXiv preprint arXiv: 2212.01878.
[37] H. Xue, S. Inati, T.S.
Sørensen, P. Kellman, M.S. Hansen, Distributed MRI
reconstruction using gadgetron-based cloud computing,
Magn. Reson. Med. 73 (3) (2015) 1015–1025.
[38] S. Mori, D. Wu, C.
Ceritoglu, Y. Li, A. Kolasny, M.A. Vaillant, A.V. Faria, K.
Oishi, M.I. Miller, MRICloud: delivering high-throughput MRI neuroinformatics as cloud-based software as a
service,
Comput. Sci. Eng. 18 (5) (2016) 21–35.
[39] F. Milletari, J. Frei, M.
Aboulatta, G. Vivar, S.-A. Ahmadi, Cloud deployment of
high-resolution medical image analysis with TOMAAT, IEEE J. Biomed. Health
Inform. 23 (3) (2018) 969–977.
[40] C.G. Xanthis, A.H. Aletras,
CoreMRI: A high-performance, publicly available MR
simulation platform on the cloud,
PLoS One 14 (5) (2019) e0216594.
[41] H. Wang, L. Tan, H.-F.
Wang, Y. Liu, R.-H. Yin, W.-Y. Wang, X.-L. Chang,
T. Jiang, J.-T. Yu, Magnetic resonance spectroscopy in Alzheimer’s disease: Systematic review and meta-analysis,
J. Alzheimers Dis. 46 (4) (2015) 1049–1070.
[42] A. Kherchouche, O.
Ben-Ahmed, C. Guillevin, B. Tremblais, A. Julian, C.
Fernandez-Maloigne, R. Guillevin, Attention-guided neural network for early dementia detection using MRS data,
Comput. Med. Imaging Graph. (2022) 102074.
[43] P. Lazen, P.L. Cardoso, S.
Sharma, C. Cadrien, T. Roetzer-Pejrimovsky, J. Furtner,
B. Strasser, L. Hingerl, A. Lipka, M. Preusser, W. Marik, W. Bogner, G. Widhalm,
K. Rössler, S. Trattnig, G. Hangel, A comparison of 7 Tesla MR spectroscopic
imaging and 3 Tesla MR fingerprinting for tumor localization in glioma patients,
2023, arXiv preprint arXiv:2304.05254.
[44] S. Nakae, K. Murayama, H.
Sasaki, M. Kumon, Y. Nishiyama, S. Ohba, K.
Adachi, S. Nagahisa, T. Hayashi, J. Inamasu, et al., Prediction of genetic subgroups in adult supra tentorial
gliomas by pre-and intraoperative parameters,
J. Neurooncol. 131 (2017) 403–412.
[45] T. Kazda, M. Bulik, P.
Pospisil, R. Lakomy, M. Smrcka, P. Slampa, R. Jancalek,
Advanced MRI increases the diagnostic accuracy of recurrent glioblastoma: Single institution thresholds and
validation of MR spectroscopy and diffusion weighted MR imaging,
NeuroImage Clin. 11 (2016) 316–321.
[46] L. Mazuel, C. Chassain, B.
Jean, B. Pereira, A. Cladière, C. Speziale, F. Durif,
Proton MR spectroscopy for diagnosis and evaluation of treatment efficacy in Parkinson disease,
Radiology 278 (2) (2016) 505–513.
[47] A. Flamez, W. Wiels, P. Van
Schuerbeek, J. De Mey, J. De Keyser, C. Baeken,
The influence of one session of low frequency rTMS on pre-supplementary motor area metabolites in late stage
Parkinson’s disease,
Clin. Neurophysiol. 130 (8) (2019) 1292–1298.
[48] H.-Y. Kim, Statistical
notes for clinical researchers: the independent samples
t-test,
Restor. Dent. Endod. 44 (3) (2019).
[49] M. Terpstra, I. Cheong, T.
Lyu, D.K. Deelchand, U.E. Emir, P. Bednařík, L.E.
Eberly, G. Öz, Test-retest reproducibility of neurochemical profiles with shortecho, single-voxel MR
spectroscopy at 3T and 7T,
Magn. Reson. Med. 76 (4) (2016) 1083–1091.
[50] I. Tkáč, G. Öz, G. Adriany,
K. Uğurbil, R. Gruetter, In vivo 1H NMR spectroscopy
of the human brain at high magnetic fields: Metabolite quantification at 4T vs. 7T,
Magn. Reson. Med. 62 (4) (2009) 868–879.
[51] V. Govindaraju, K. Young,
A.A. Maudsley, Proton NMR chemical shifts and
coupling constants for brain metabolites,
NMR Biomed. 13 (3) (2000) 129–153.
[52] R. Kreis, The trouble with
quality filtering based on relative C ramér-R ao lower
bounds,
Magn. Reson. Med. 75 (1) (2016) 15–18.
[53] A. Metastasio, P. Rinaldi,
R. Tarducci, E. Mariani, F.T. Feliziani, A. Cherubini,
G.P. Pelliccioli, G. Gobbi, U. Senin, P. Mecocci, Conversion of MCI to dementia: Role of proton magnetic
resonance spectroscopy,
Neurobiol. Aging 27 (7) (2006) 926–932.
[54] K. Kantarci, Proton MRS in
mild cognitive impairment,
J. Magn. Reson. Imaging 37 (4) (2013) 770–777.
[55] S. Tumati, S. Martens, A.
Aleman, Magnetic resonance spectroscopy in mild
cognitive impairment: Systematic review and meta-analysis,
Neurosci. Biobehav. Rev. 37 (10) (2013) 2571–2586.