pFISTA-SENSE-ResNet for Parallel MRI Reconstruction ( [English] )
路铁源1, 张心林1, 黄奕晖1, 郭迪2, 黄峰3, 许勤3, 胡榆涵1, 欧阳林4,5, 林建忠6, 颜志平
7, 屈小波1**
1厦门大学,电子科学系,福建等离子体与磁共振重点研究实验室,中国,厦门;
2厦门理工学院,计算机与信息工程学院,中国,厦门;
3东软医疗系统有限公司,中国,上海;;
4厦门大学医学院附属东南医院医学影像科,中国,漳州;.
5厦门大学医学院医学影像研究所,中国,漳州;;
6厦门大学附属中山医院磁共振科,中国,厦门;.
7厦门市弘爱医院放射科,中国,厦门..
联系人:
quxiaobo<|at|>xmu.edu.cn
引用: Tieyuan Lu, Xinlin Zhang, Yihui Huang, Di Guo, Feng Huang, Qin Xu, Yuhan Hu, Lin Ou-Yang, Jianzhong Lin, Zhiping Yan, Xiaobo Qu*, pFISTA-SENSE-ResNet for Parallel MRI Reconstruction,Journal of Magnetic Resonance, DOI: 10.1016/j.jmr.2020.106790, 2020.
全文链接:https://authors.elsevier.com/a/1bW4K3u0yjN8GV
摘要:
虽然磁共振成像(Magnetic resonance imaging, MRI)已经被广泛应用于临床诊断中,但仍受制于其较长的采集时间。尽管可以通过稀疏采样和并行成像来加速成像过程,但是以较快的计算速度获得令人满意的图像仍然充满了挑战。最近深度学习的方法凭借其令人鼓舞的重建结果备受关注,但它们在理论上缺乏一定的可解释性。在本文的工作中,我们从稀疏迭代重建的角度来设计网络结构,并使用残差结构来增强网络的性能,以此来确保磁共振并行成像的高质量重建结果。公开的膝盖数据的实验结果表明,与最先进的深度学习方法和最优化方法相比,所提方法重建的图像误差更低,对采样模板也更为鲁棒.
关键词:
磁共振成像,图像重建,深度学习,稀疏学习,网络可解释性。
方法:
1.
背景
更好的图像质量和更快的重建速度是MRI重建中的关键问题,充满挑战的同时也非常值得研究。为了能够获得更低的重建误差,压缩感知(compressed sensing, CS)方法中已经使用了包括固定的和自适应的稀疏变换。近年来,基于数据集的深度学习方法被应用于许多领域,包括生物磁共振波谱,医学图像分析以及加速MRI图像重建,并凭借强大的深度卷积神经网络(convolution neural network,CNN)表现出了优异的性能。然而,与基于最优化的迭代算法相比,CNN在图像重建的过程中像是一个黑盒子,缺乏可解释性。
为了提高网络的可解释性,一些最优化算法被展开为了深度学习网络,比如:VN和MoDL。但是,VN中使用单个卷积层作为正则项限制了其重建质量;MoDL将共轭梯度算法引入网络来解决数据校验的子问题,这降低了它的重建速度。
2.
pFISTA-SENSE-ResNet
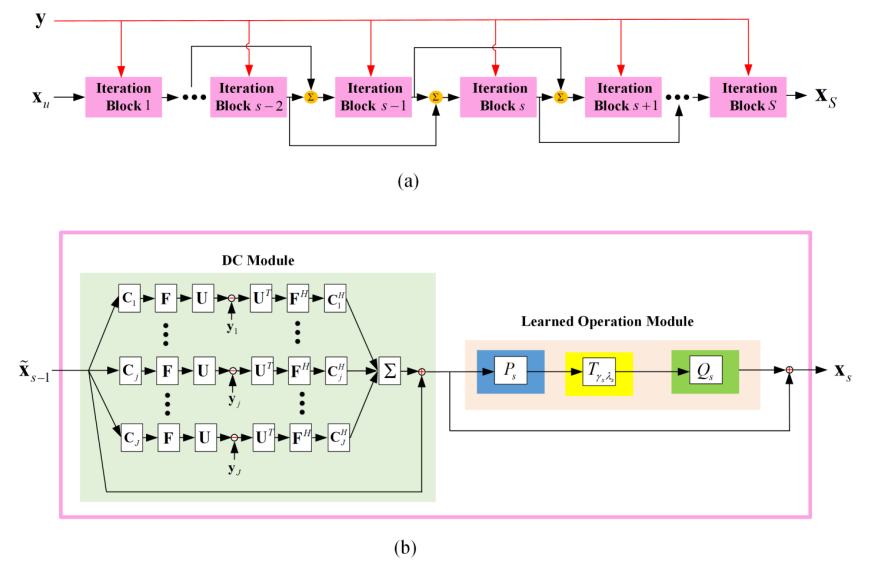
我们将pFISTA-SENSE 的展开形式作为网络的基本结构,网络的第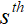 个迭代块可以表示为:
个迭代块可以表示为:
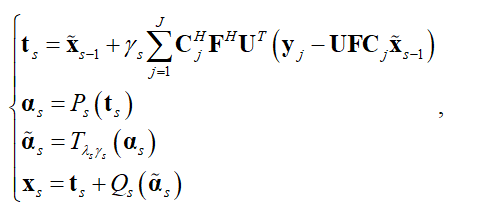
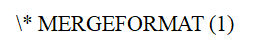
其中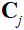 是第
是第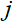 个线圈的灵敏度地图,
个线圈的灵敏度地图,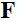 是傅里叶变换,
是傅里叶变换, 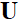 是欠采样矩阵,
是欠采样矩阵,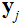 是第
是第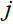 个线圈采样到的k空间数据,
个线圈采样到的k空间数据,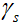 是步长,
是步长,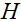 和
和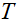 分别表示共轭转置和转置,
分别表示共轭转置和转置,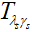 是软阈值函数。
是软阈值函数。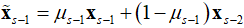 。
。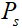 和
和 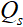 是由CNN组成的前向操作和反向操作。
是由CNN组成的前向操作和反向操作。
本文所提的网络结构如图1所示。
3.
主要结果
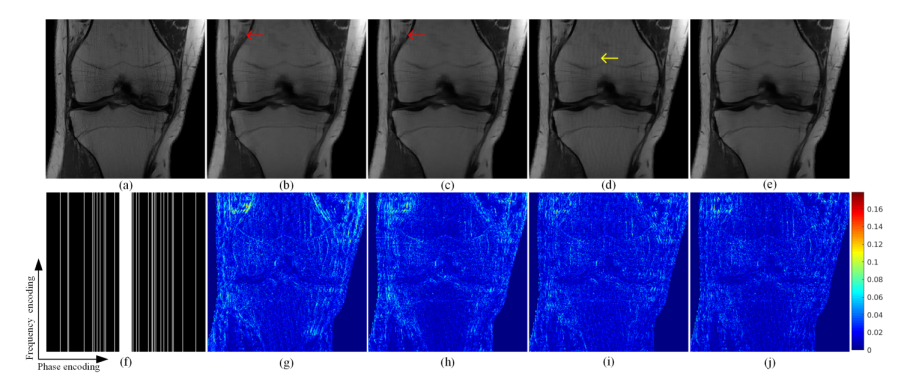
本文所提方法与基于最优化的pFISTA-SENSE和两种深度学习方法:VN以及MoDL进行了实验对比。图2展示了公开膝盖数据集7倍加速下各个方法的重建结果。与其他方法相比,pFISTA-SENSE-ResNet的重建结果最接近与全采样图像。首先,pFISTA-SENSE-ResNet 的重建结果(图2(e))比pFISTA-SENSE的重建结果(图2(b))更清晰,对欠采样伪影的抑制也要比MoDL的重建结果(图2(d))好,如图2(d)中黄色箭头所指。其次,小细节也有更好的重建出来,如图2中红色箭头所指。除此以外,根据误差图来看,pFISTA-SENSE-ResNet重建的误差要更低。
致谢:
这项工作得到了国家重点研发计划(2017YFC0108703),国家自然科学基金(61971361、61871341和61811530021),福建省自然科学基金(2018J06018),中央高校基本科研基金(20720180056)和厦门大学南强杰出人才计划的自处。作者要感谢英伟达公司捐赠的GPU,作者还要感谢厦门医学院附属第二医院放射影像科的杨永贵和郭岗,以及厦门大学电子科学系包立君对本文的修改。最后,作者要感谢审稿人对本文提出的宝贵意见。
参考文献:
[1] M. Lustig, D. Donoho, and J. M. Pauly, "Sparse MRI: The application of compressed sensing for rapid MR imaging," Magnetic Resonance in Medicine, vol. 58, no. 6, pp. 1182-1195, 2007.
[2] J. Hamilton, D. Franson, and N. Seiberlich, "Recent advances in parallel imaging for MRI," Progress in Nuclear Magnetic Resonance Spectroscopy, vol. 101, pp. 71-95, 2017.
[3] Z.-P. Liang, "Spatiotemporal imaging with partially separable functions," in 2007 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2007, pp. 988-991.
[4] X. Qu, Y. Hou, F. Lam, D. Guo, J. Zhong, and Z. Chen, "Magnetic resonance image reconstruction from undersampled measurements using a patch-based nonlocal operator," Medical Image Analysis, vol. 18, no. 6, pp. 843-856, 2014.
[5] K. P. Pruessmann, M. Weiger, M. B. Scheidegger, and P. Boesiger, "SENSE: Sensitivity encoding for fast MRI," Magnetic Resonance in Medicine, vol. 42, no. 5, pp. 952-962, 1999.
[6] J. Jin, F. Liu, E. Weber, Y. Li, and S. Crozier, "An electromagnetic reverse method of coil sensitivity mapping for parallel MRI–Theoretical framework," Journal of Magnetic Resonance, vol. 207, no. 1, pp. 59-68, 2010.
[7] B. Liu, F. Sebert, Y. Zou, and L. Ying, "SparseSENSE: Randomly-sampled parallel imaging using compressed sensing," in 16th Annual Meeting of ISMRM, 2008, p. 3154.
[8] D. Liang, B. Liu, J. Wang, and L. Ying, "Accelerating SENSE using compressed sensing," Magnetic Resonance in Medicine, vol. 62, no. 6, pp. 1574-1584, 2009.
[9] X. Qu, W. Zhang, D. Guo, C. Cai, S. Cai, and Z. Chen, "Iterative thresholding compressed sensing MRI based on contourlet transform," Inverse Problems in Science and Engineering, vol. 18, no. 6, pp. 737-758, 2010.
[10] S. Pejoski, V. Kafedziski, and D. Gleich, "Compressed sensing MRI using discrete nonseparable shearlet transform and FISTA," IEEE Signal Processing Letters, vol. 22, no. 10, pp. 1566-1570, 2015.
[11] X. Qu et al., "Undersampled MRI reconstruction with patch-based directional wavelets," Magnetic Resonance Imaging, vol. 30, no. 7, pp. 964-977, 2012.
[12] S. Ravishankar and Y. Bresler, "MR image reconstruction from highly undersampled k-space data by dictionary learning," IEEE Transactions on Medical Imaging, vol. 30, no. 5, pp. 1028-1041, 2010.
[13] Z. Lai et al., "Image reconstruction of compressed sensing MRI using graph-based redundant wavelet transform," Medical Image Analysis, vol. 27, pp. 93-104, 2016.
[14] Z. Zhan, J.-F. Cai, D. Guo, Y. Liu, Z. Chen, and X. Qu, "Fast multiclass dictionaries learning with geometrical directions in MRI reconstruction," IEEE Transactions on Biomedical Engineering, vol. 63, no. 9, pp. 1850-1861, 2015.
[15] X. Qu et al., "Accelerated nuclear magnetic resonance spectroscopy with deep learning," Angewandte Chemie International Edition, vol. 59, no. 26, pp. 10297-10300, 2020.
[16] D. Chen, Z. Wang, D. Guo, V. Orekhov, and X. Qu, "Review and prospect: Deep learning in nuclear magnetic resonance spectroscopy," Chemistry-A European Journal, 2020. DOI: 10.1002/chem.202000246.
[17] W. Lin et al., "Convolutional neural networks-based MRI image analysis for the alzheimer’s disease prediction from mild cognitive impairment," Frontiers in Neuroscience, vol. 12, p. 777, 2018.
[18] S. Wang et al., "Accelerating magnetic resonance imaging via deep learning," in IEEE International Symposium on Biomedical Imaging, 2016, pp. 514-517.
[19] J. Zhang et al., "Robust single-shot T 2 mapping via multiple overlapping-echo acquisition and deep neural network," IEEE Transactions on Medical Imaging, vol. 38, no. 8, pp. 1801-1811, 2019.
[20] L. Bao et al., "Undersampled MR image reconstruction using an enhanced recursive residual network," Journal of Magnetic Resonance, vol. 305, pp. 232-246, 2019.
[21] J. Schlemper, J. Caballero, J. V. Hajnal, A. N. Price, and D. Rueckert, "A deep cascade of convolutional neural networks for dynamic MR image reconstruction," IEEE Transactions on Medical Imaging, vol. 37, no. 2, pp. 491-503, 2018.
[22] K. Zeng, Y. Yang, G. Xiao, and Z. Chen, "A very deep densely connected network for compressed sensing MRI," IEEE Access, vol. 7, pp. 85430-85439, 2019.
[23] Y. Yang, J. Sun, H. Li, and Z. Xu, "Deep ADMM-Net for compressive sensing MRI," in Advances in Neural Information Processing Systems, 2016, pp. 10-18.
[24] J. Zhang and B. Ghanem, "ISTA-Net: Interpretable optimization-inspired deep network for image compressive sensing," in IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 1828-1837.
[25] J. Cheng, H. Wang, Y. Zhu, Q. Liu, L. Ying, and D. Liang, "Model-based deep MR imaging: The roadmap of generalizing compressed sensing model using deep learning," arXiv preprint arXiv:1906.08143, 2019.
[26] K. Hammernik et al., "Learning a variational network for reconstruction of accelerated MRI data," Magnetic Resonance in Medicine, vol. 79, no. 6, pp. 3055-3071, 2017.
[27] L. Landweber, "An iteration formula for Fredholm integral equations of the first kind," American Journal of Mathematics, vol. 73, no. 3, pp. 615-624, 1951.
[28] H. K. Aggarwal, M. P. Mani, and M. Jacob, "MoDL: Model based deep learning architecture for inverse problems," IEEE Transactions on Medical Imaging, vol. 38, no. 2, pp. 394-405, 2018.
[29] Y. Liu, Z. Zhan, J. Cai, D. Guo, Z. Chen, and X. Qu, "Projected iterative soft-thresholding algorithm for tight frames in compressed sensing magnetic resonance imaging," IEEE Transactions on Medical Imaging, vol. 35, no. 9, pp. 2130-2140, 2016.
[30] I. Daubechies, M. Defrise, and C. De Mol, "An iterative thresholding algorithm for linear inverse problems with a sparsity constraint," Communications on Pure and Applied Mathematics, vol. 57, no. 11, pp. 1413-1457, 2004.
[31] A. Beck and M. Teboulle, "A fast iterative shrinkage-thresholding algorithm for linear inverse problems," SIAM Journal on Imaging Sciences, vol. 2, no. 1, pp. 183-202, 2009.
[32] Y. Liu et al., "Balanced sparse model for tight frames in compressed sensing magnetic resonance imaging," PLoS One, vol. 10, no. 4, p. e0119584, 2015.
[33] S. T. Ting et al., "Fast implementation for compressive recovery of highly accelerated cardiac cine MRI using the balanced sparse model," Magnetic Resonance in Medicine, vol. 77, no. 4, pp. 1505-1515, 2017.
[34] X. Zhang, H. Lu, D. Guo, L. Bao, F. Huang, and X. Qu, "A convergence proof of projected fast iterative soft-thresholding algorithm for parallel magnetic resonance imaging," arXiv preprint arXiv:1909.07600, 2019.
[35] V. Nair and G. E. Hinton, "Rectified linear units improve restricted boltzmann machines," in 27th International Conference on Machine Learning, 2010, pp. 807-814.
[36] K. He, X. Zhang, S. Ren, and J. Sun, "Deep residual learning for image recognition," in IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770-778.
[37] M. Uecker et al., "ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA," Magnetic Resonance in Medicine, vol. 71, no. 3, pp. 990-1001, 2014.
[38] X. Glorot and Y. Bengio, "Understanding the difficulty of training deep feedforward neural networks," in Proceedings of the 13th International Conference on Artificial Intelligence and Statistics, 2010, pp. 249-256.
[39] D. P. Kingma and J. Ba, "Adam: A method for stochastic optimization," arXiv preprint arXiv:1412.6980, 2014.
[40] Z. Wang, A. C. Bovik, H. R. Sheikh, and E. P. Simoncelli, "Image quality assessment: From error visibility to structural similarity," IEEE Transactions on Image Processing, vol. 13, no. 4, pp. 600-612, 2004.
[41] D. Narnhofer, K. Hammernik, F. Knoll, and T. Pock, "Inverse GANs for accelerated MRI reconstruction," in Wavelets and Sparsity XVIII, 2019.
[42] Y. Han, J. Yoo, H. H. Kim, H. J. Shin, and J. C. Ye, "Deep learning with domain adaptation for accelerated projection-reconstruction MR," Magnetic Resonance in Medicine, vol. 80, no. 3, pp. 1189-1205, 2018.